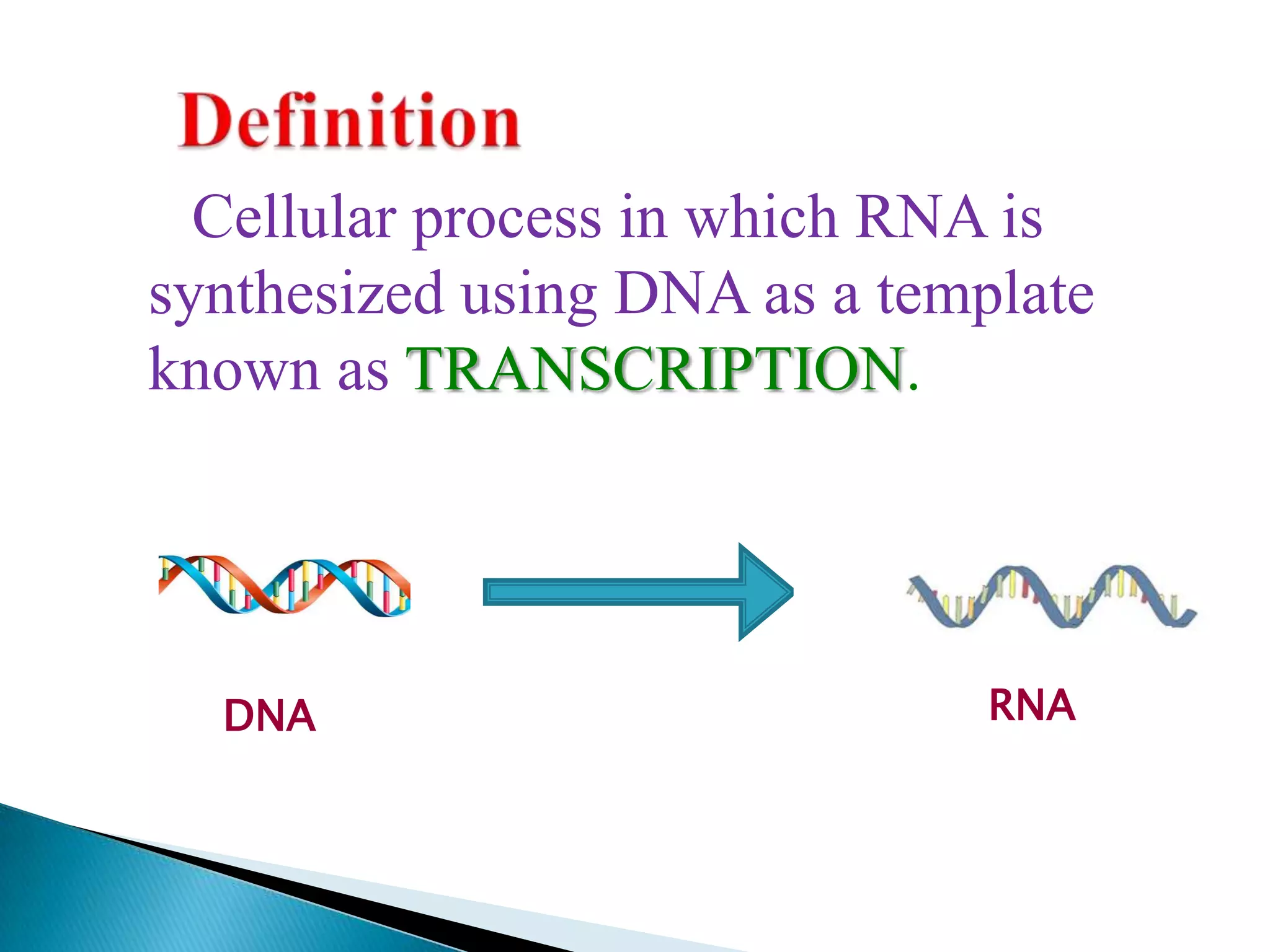
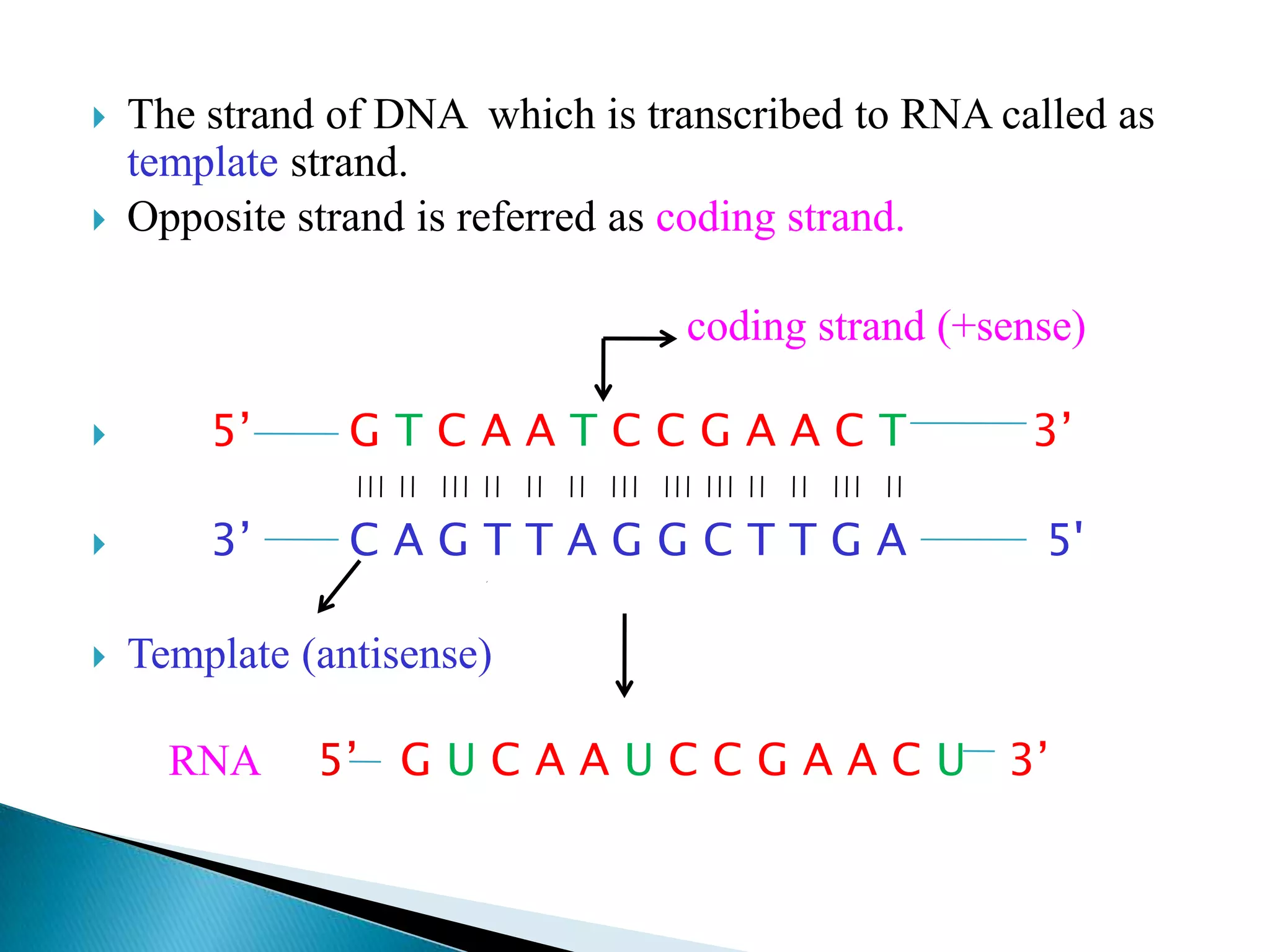
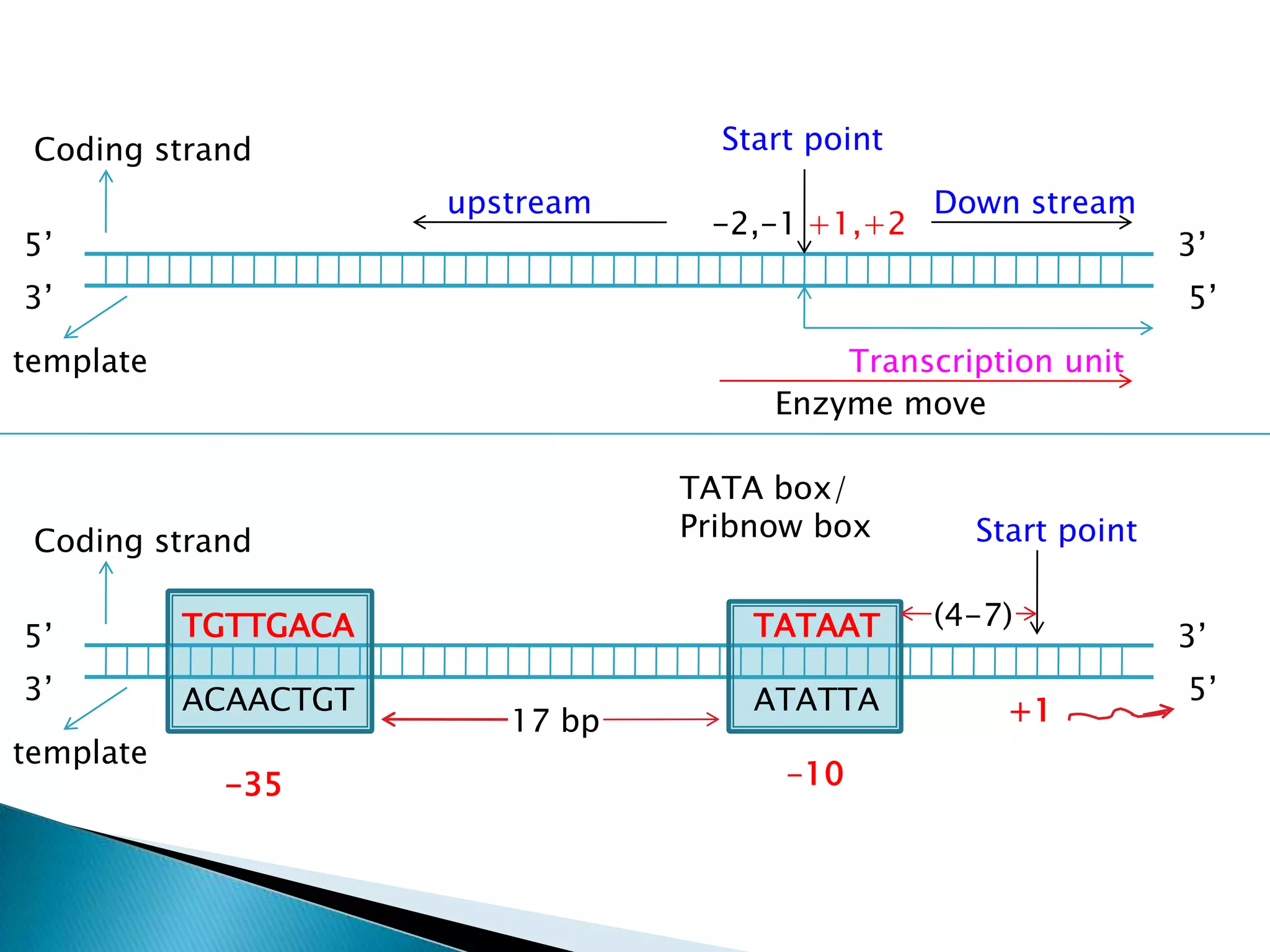
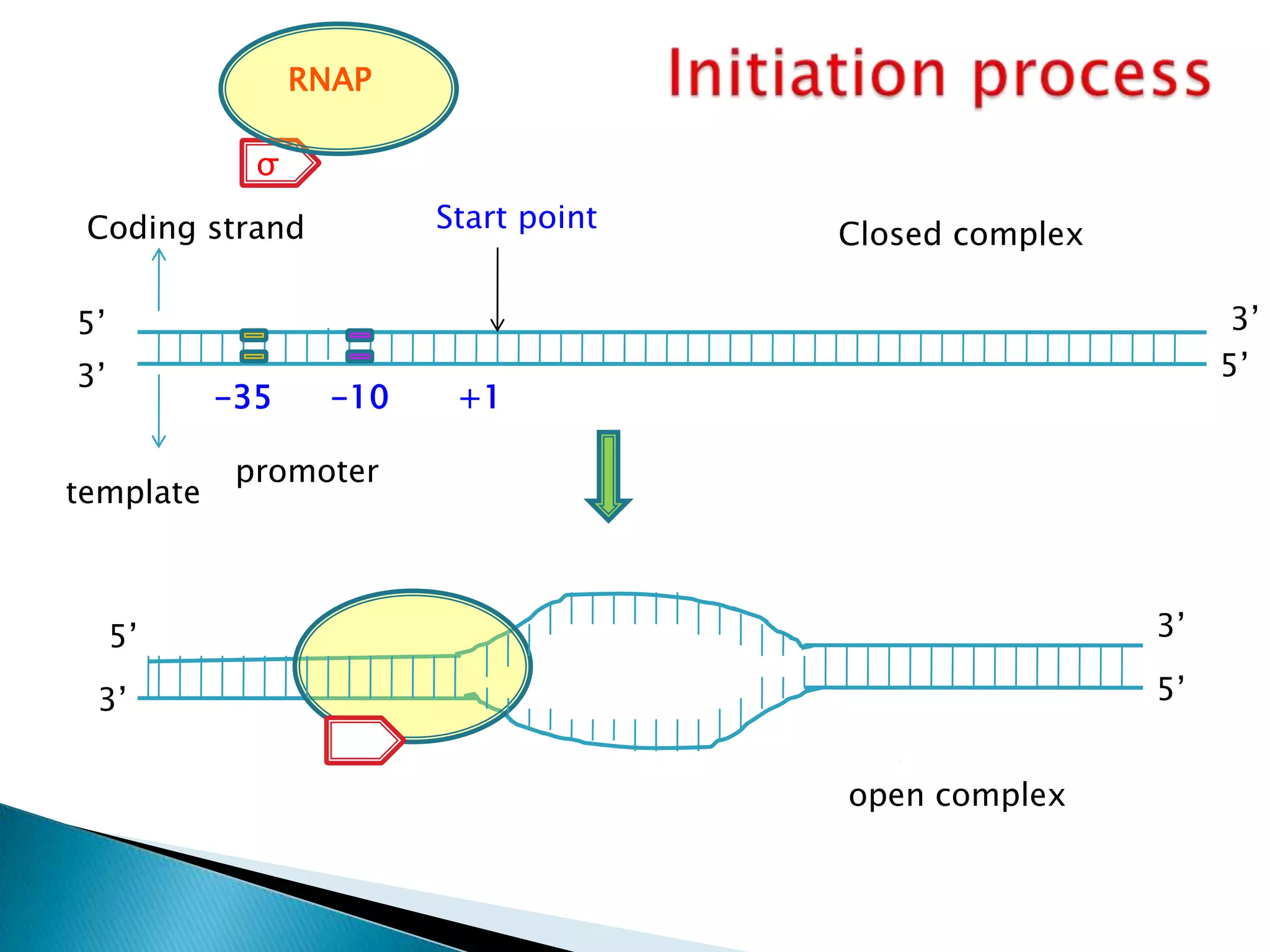
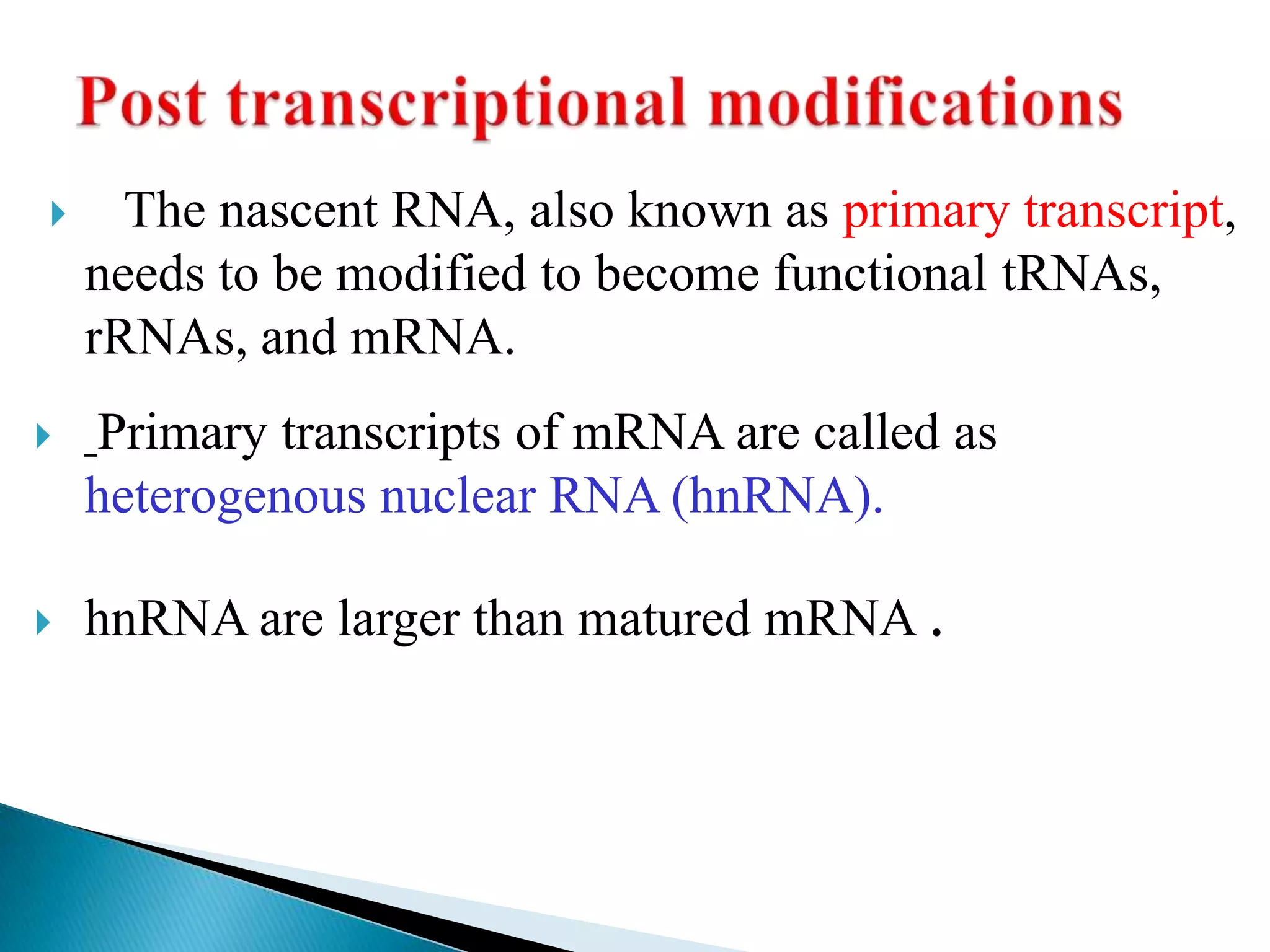
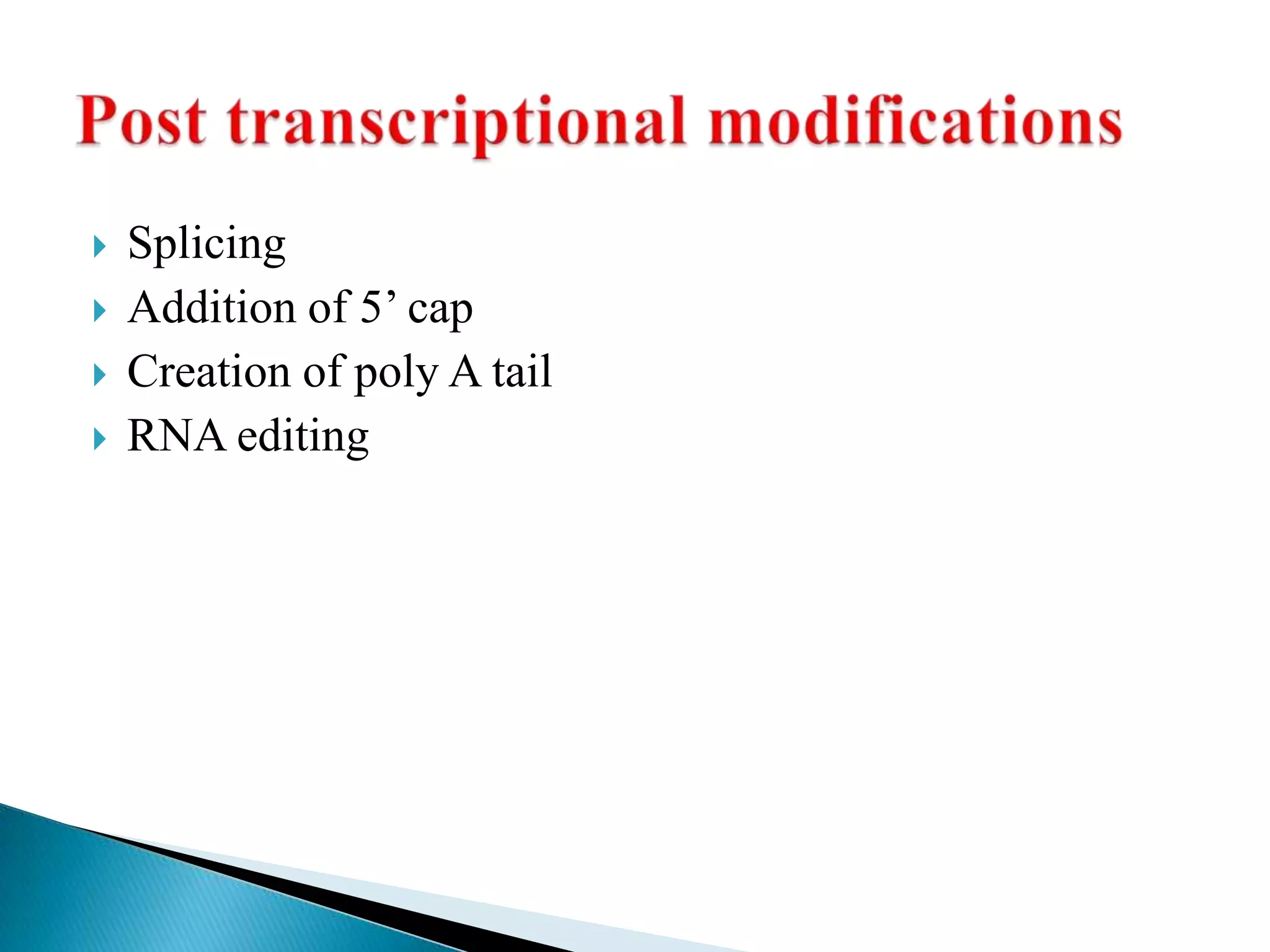
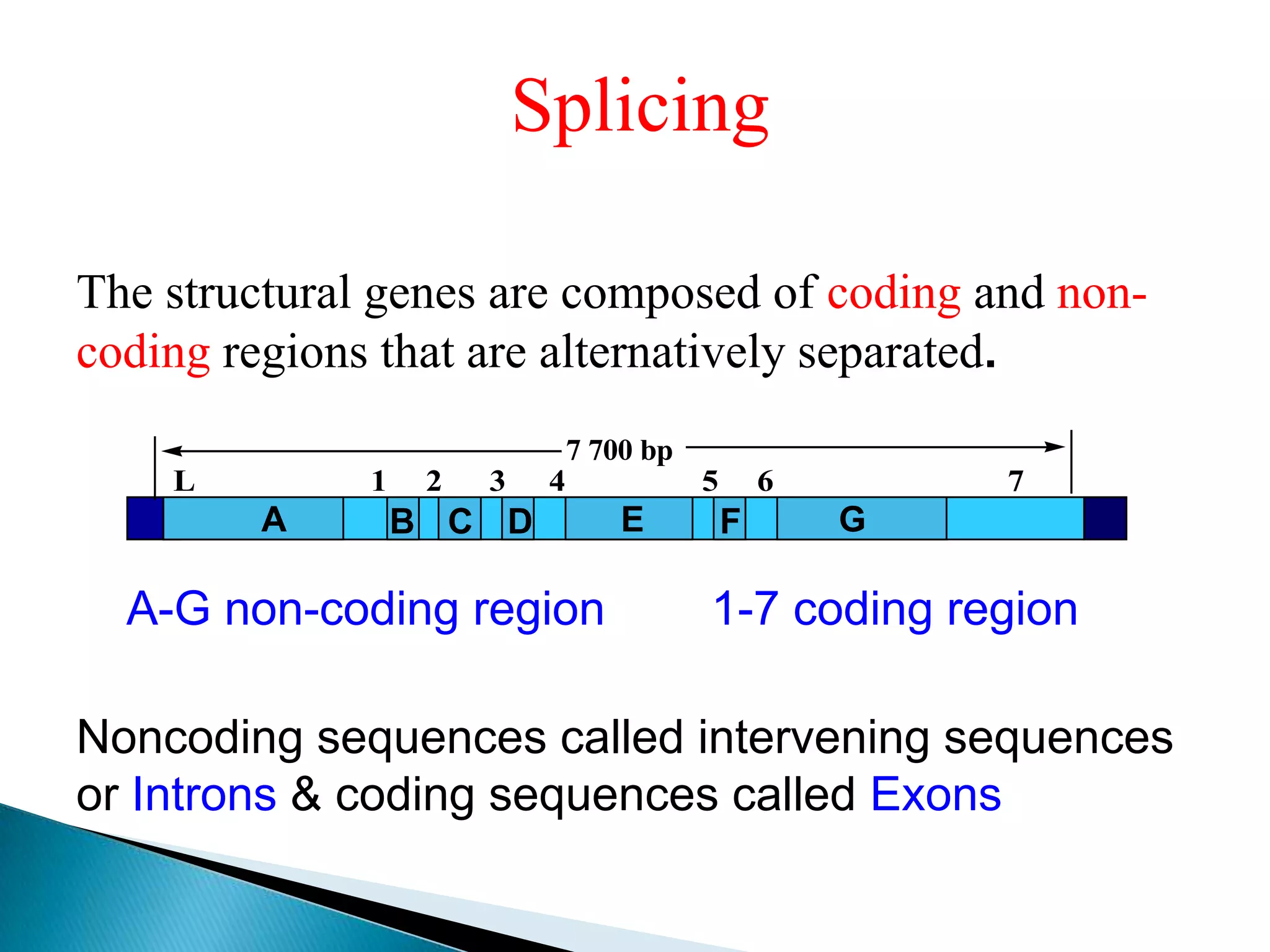
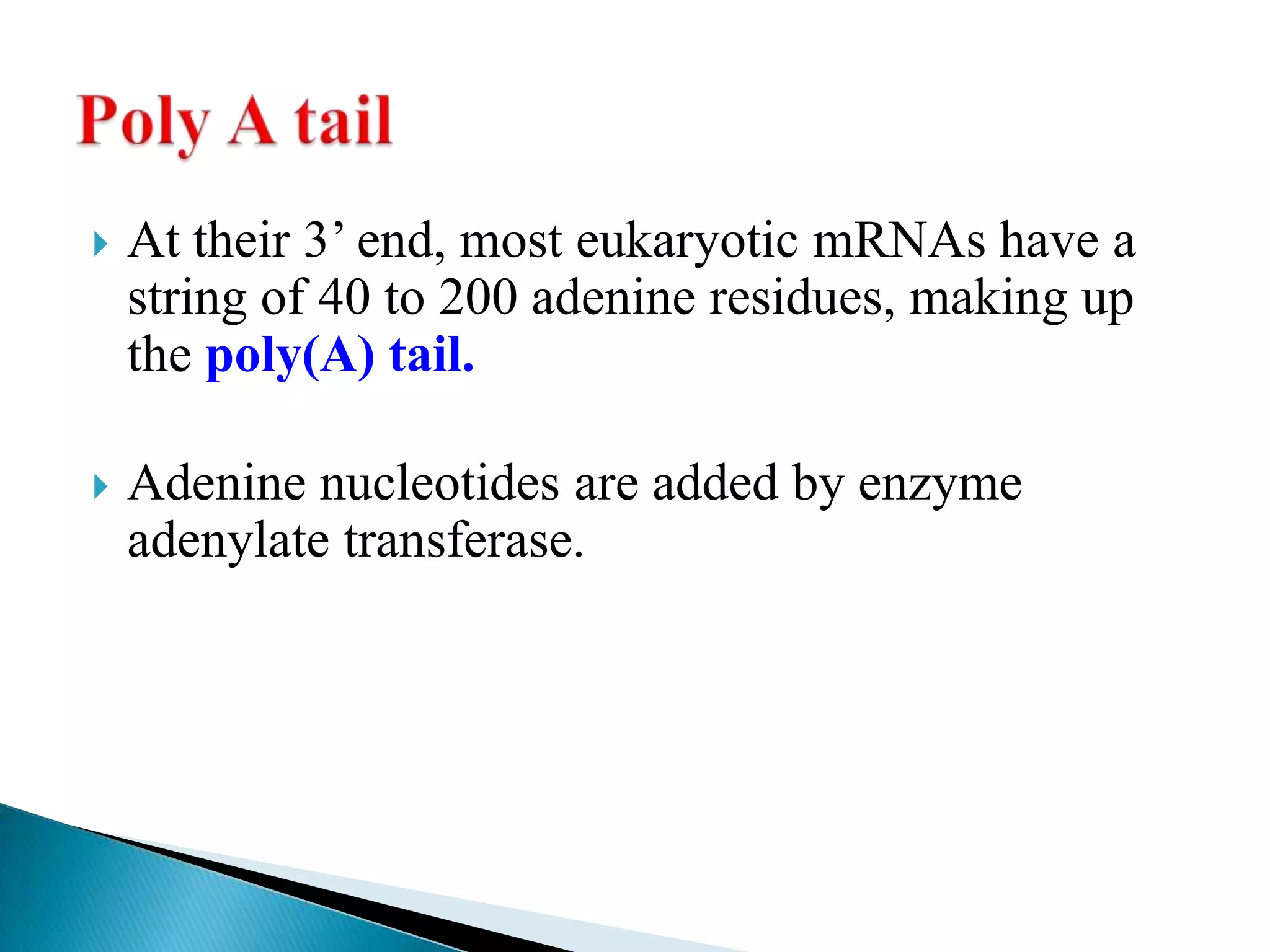
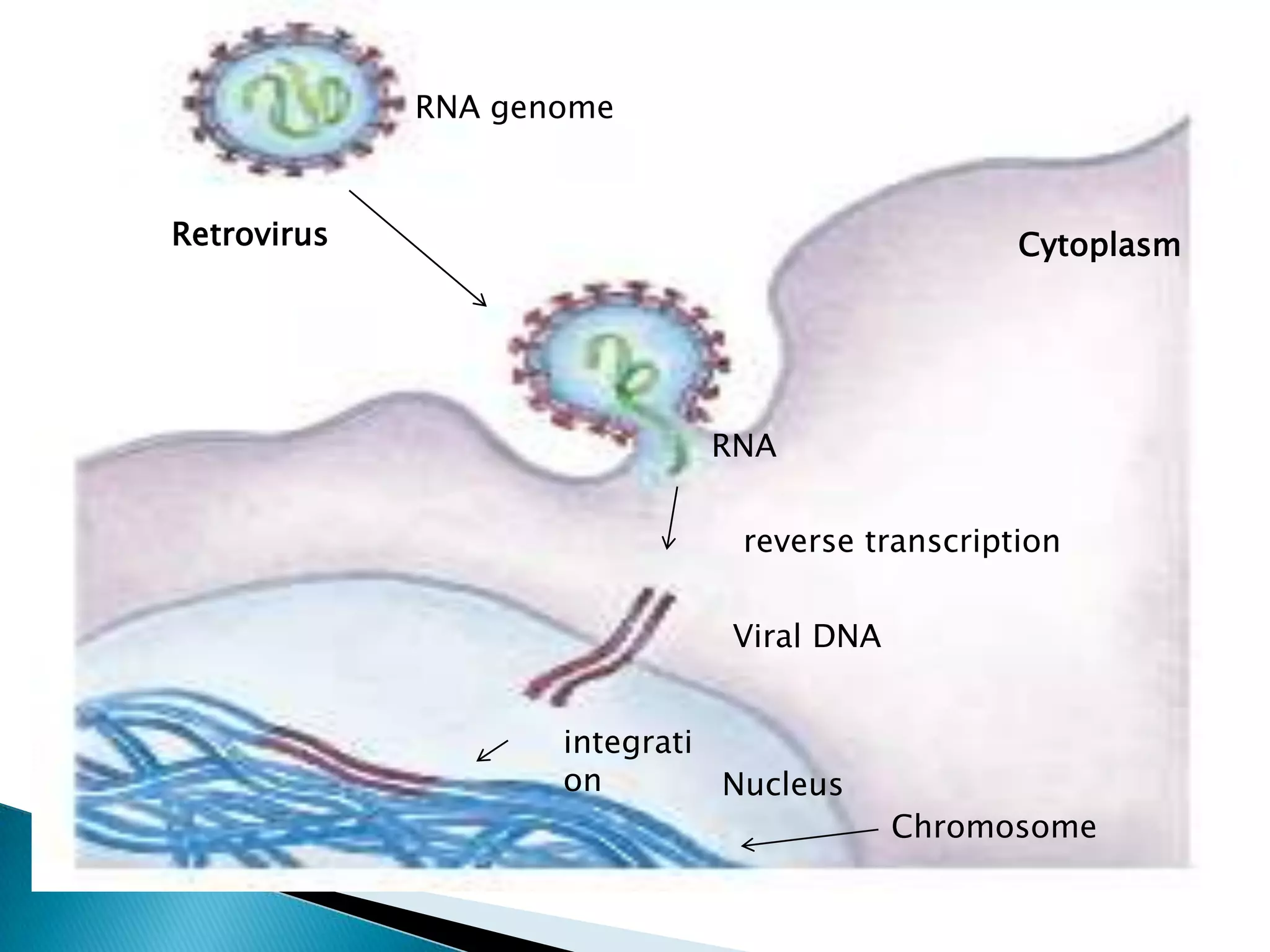
1. The document discusses transcription, the process by which RNA is synthesized using DNA as a template.
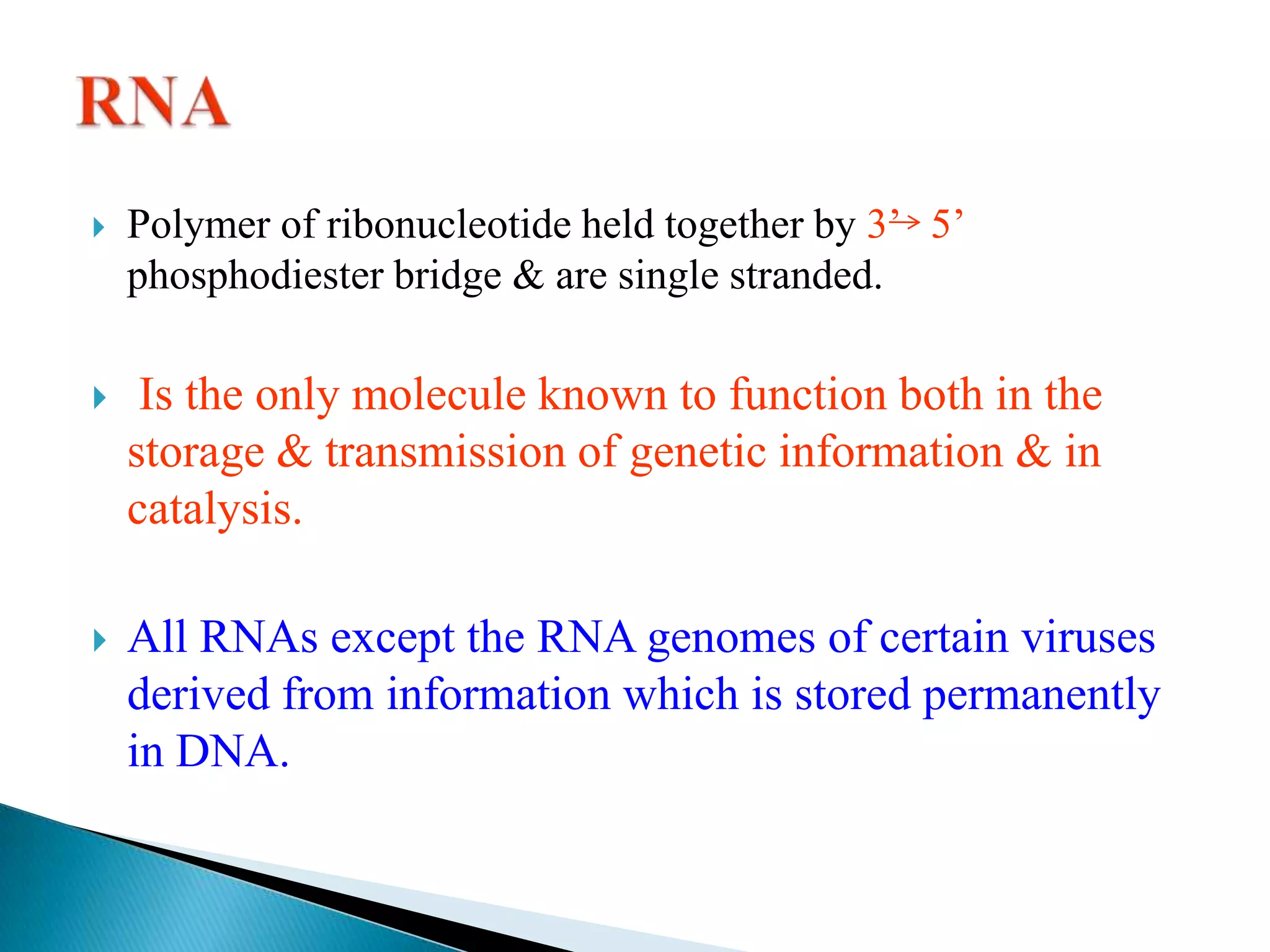
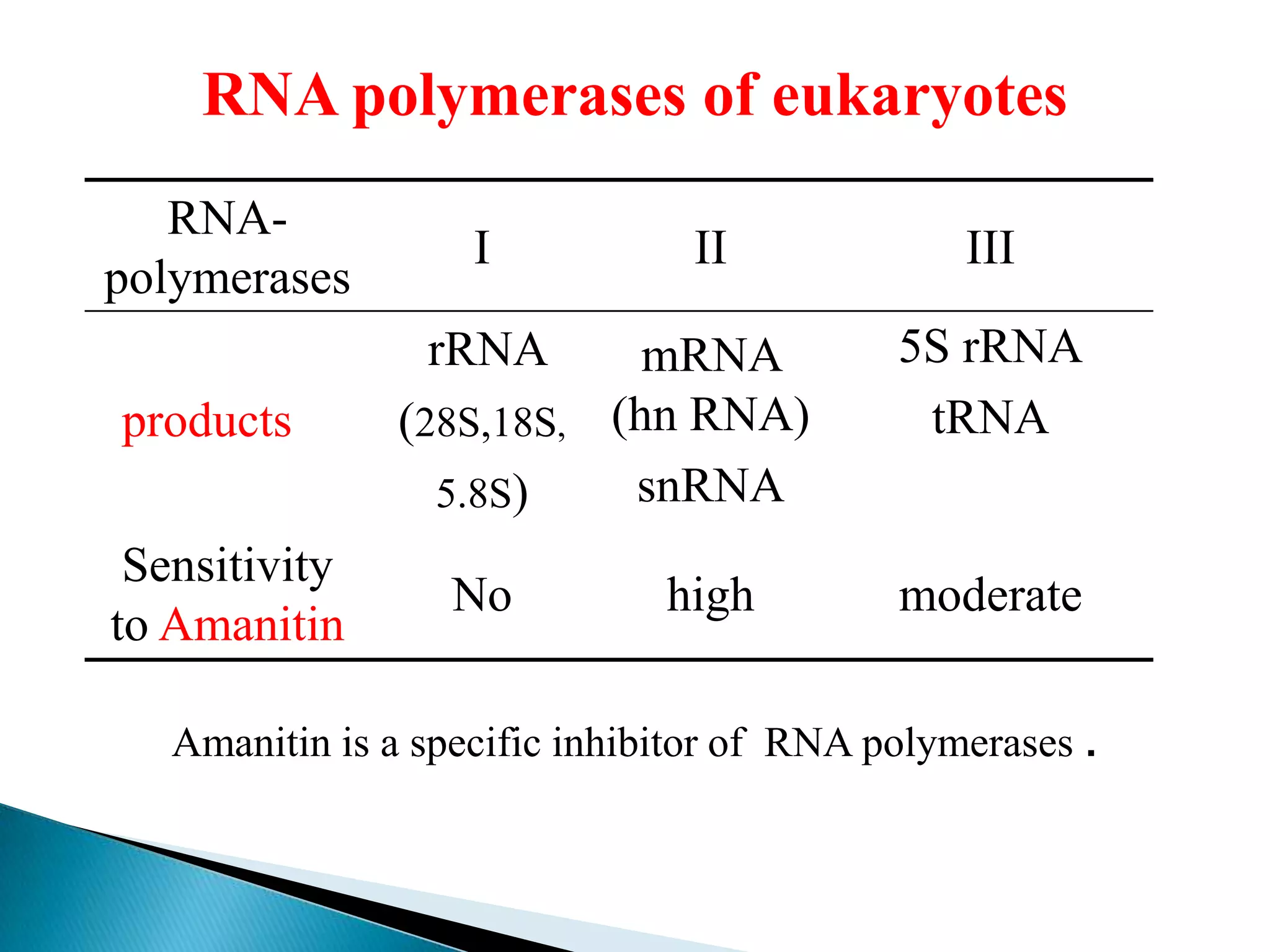
2. There are three main types of RNA - mRNA, tRNA, and rRNA - which have different functions like encoding proteins, transporting amino acids, and constituting ribosomes.
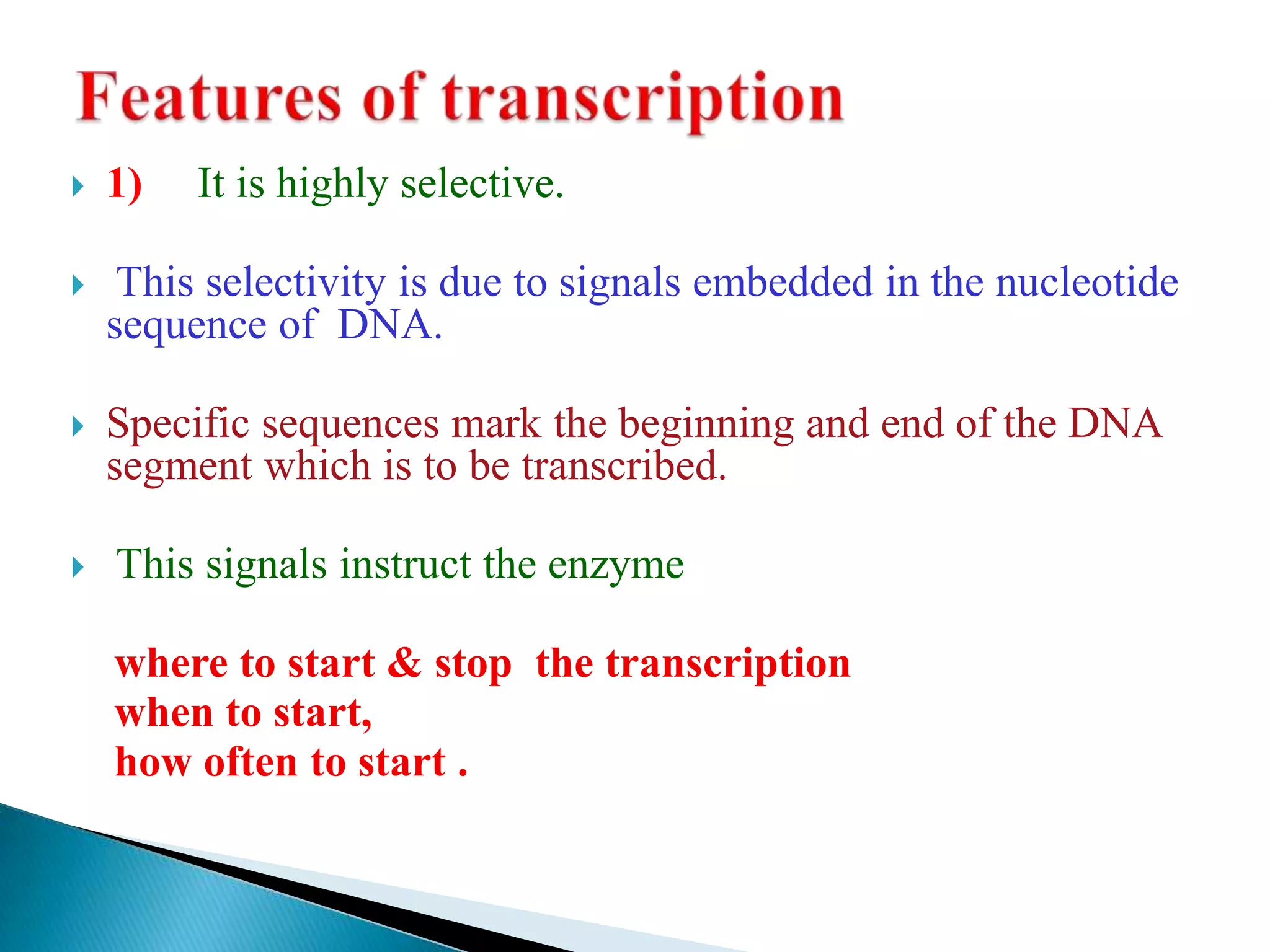
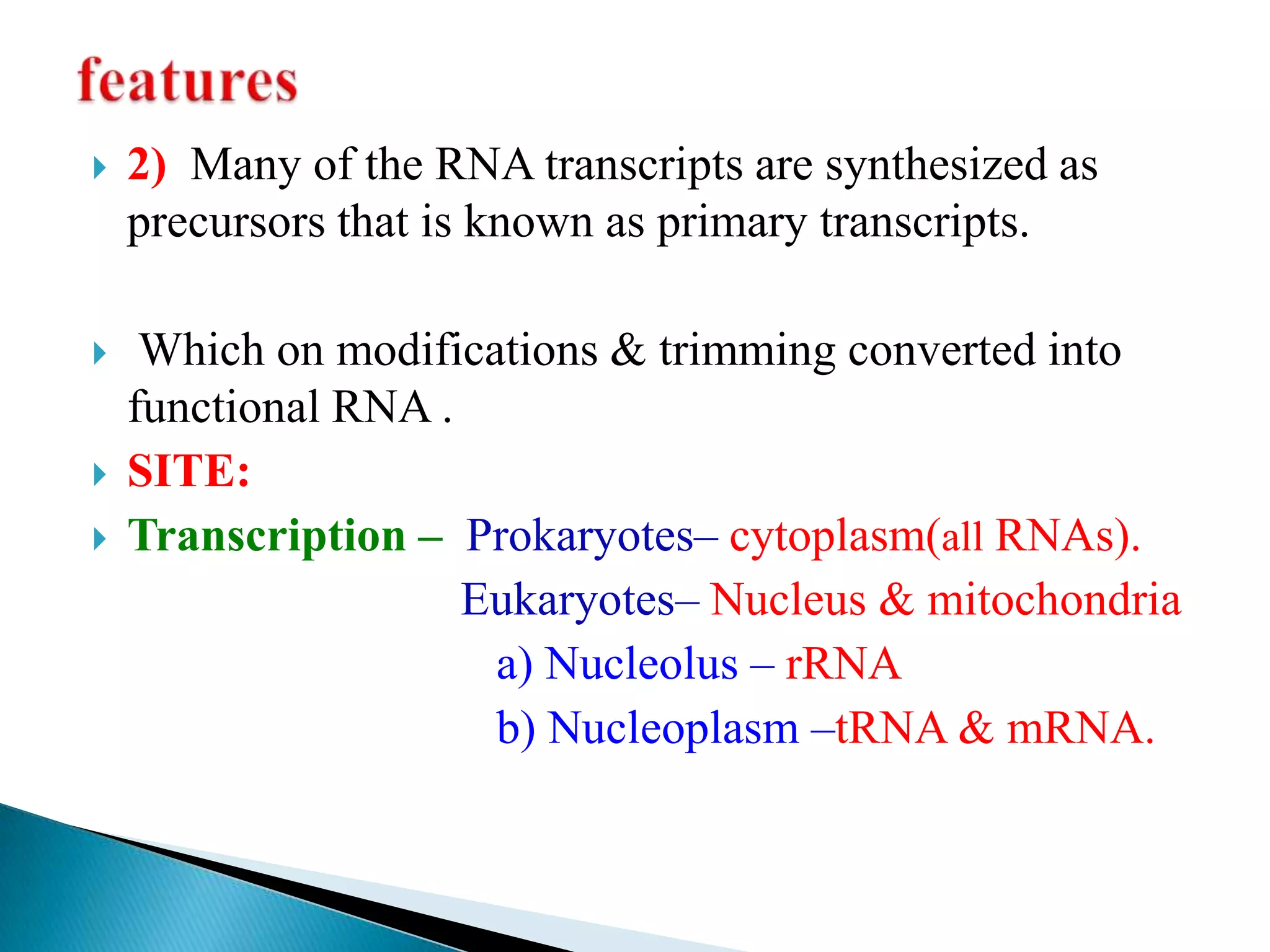
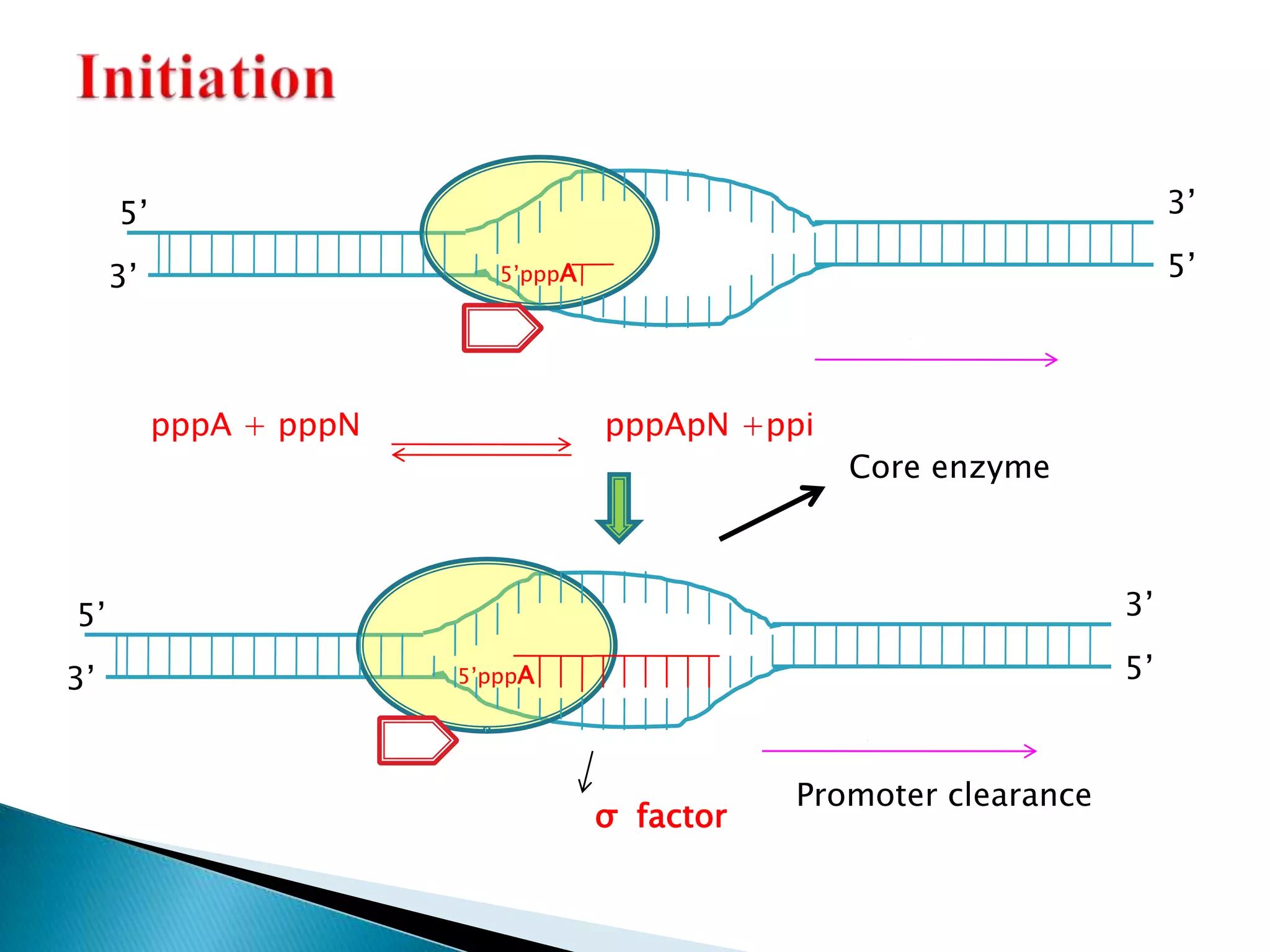
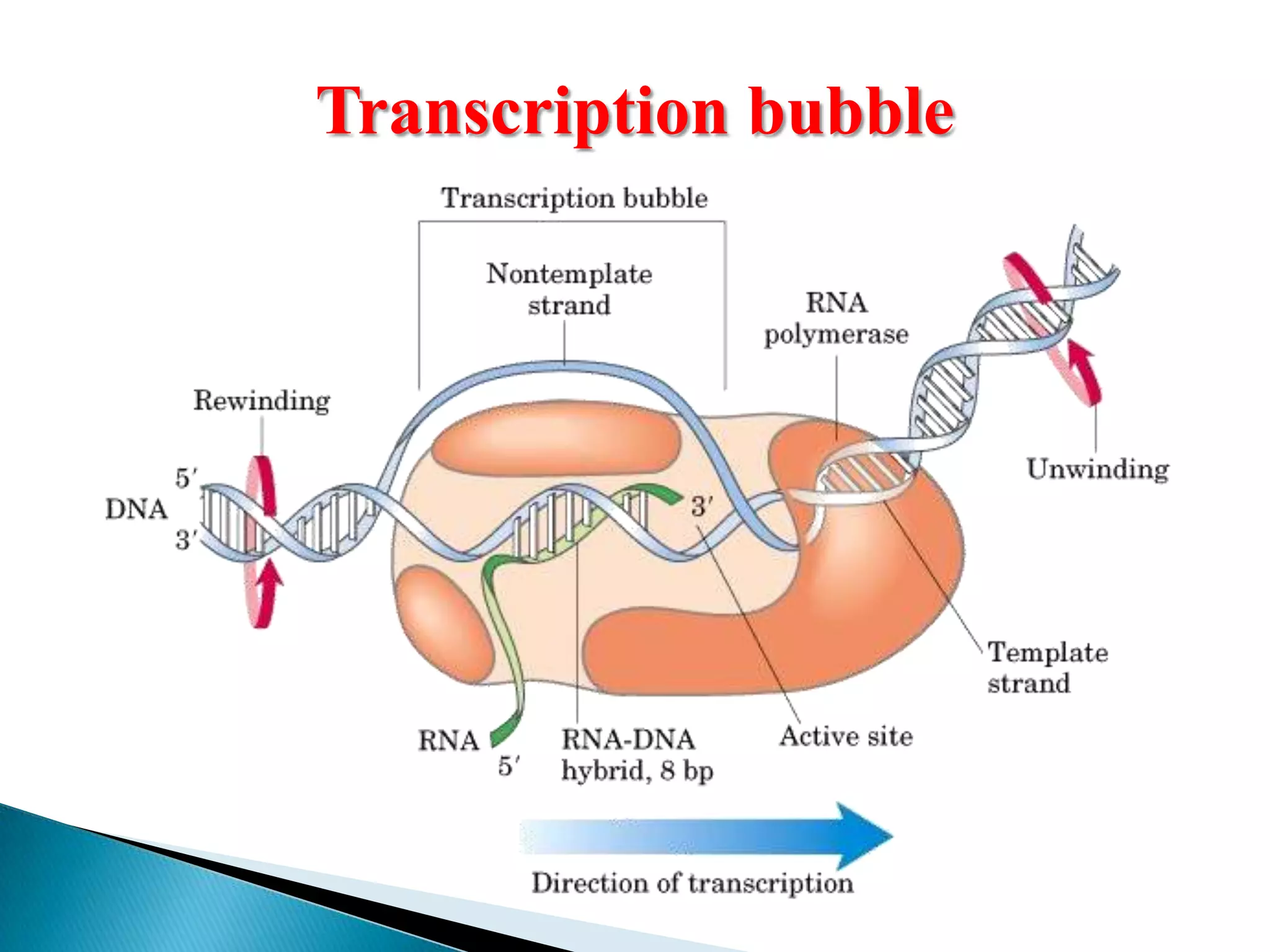
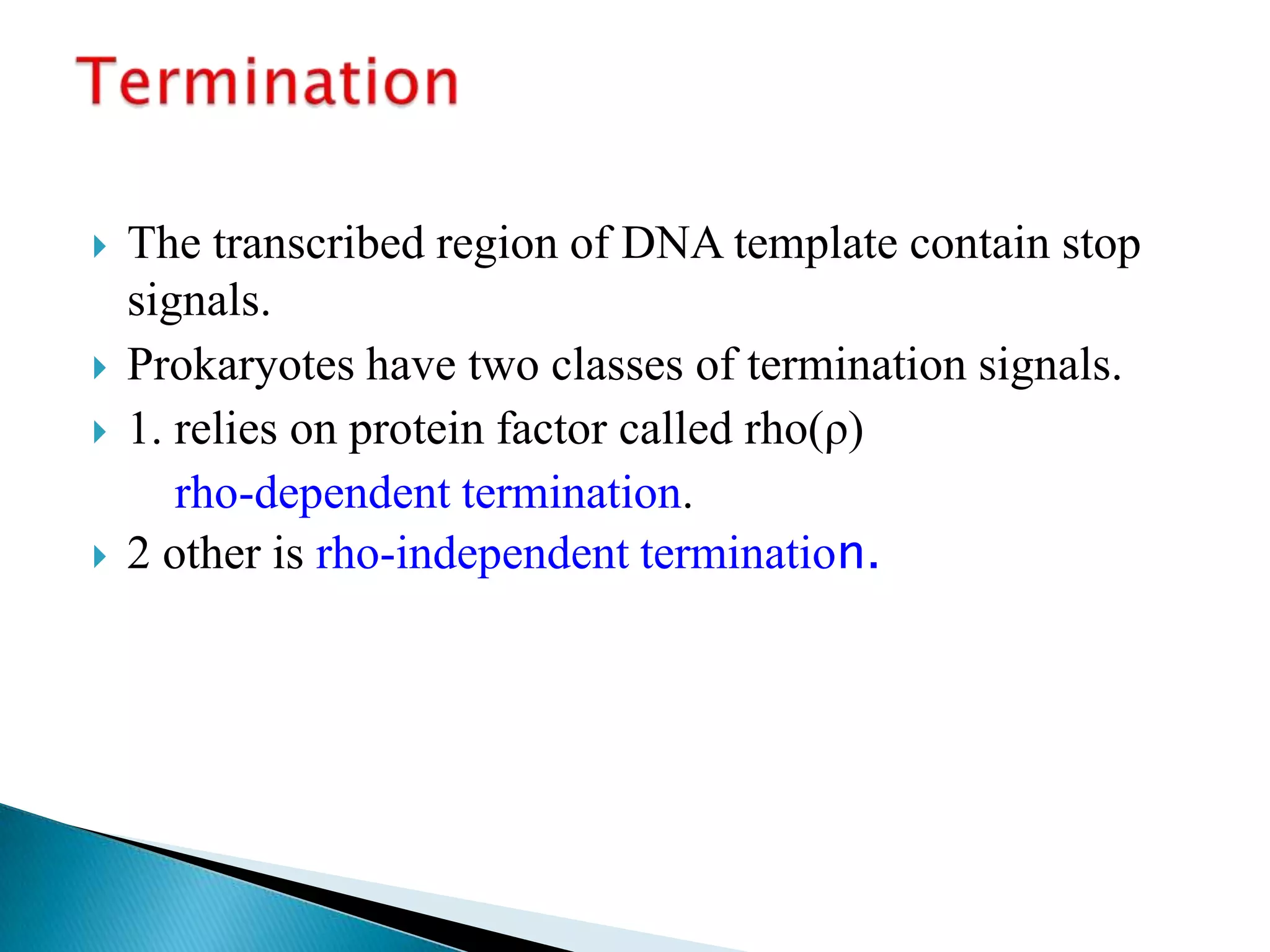
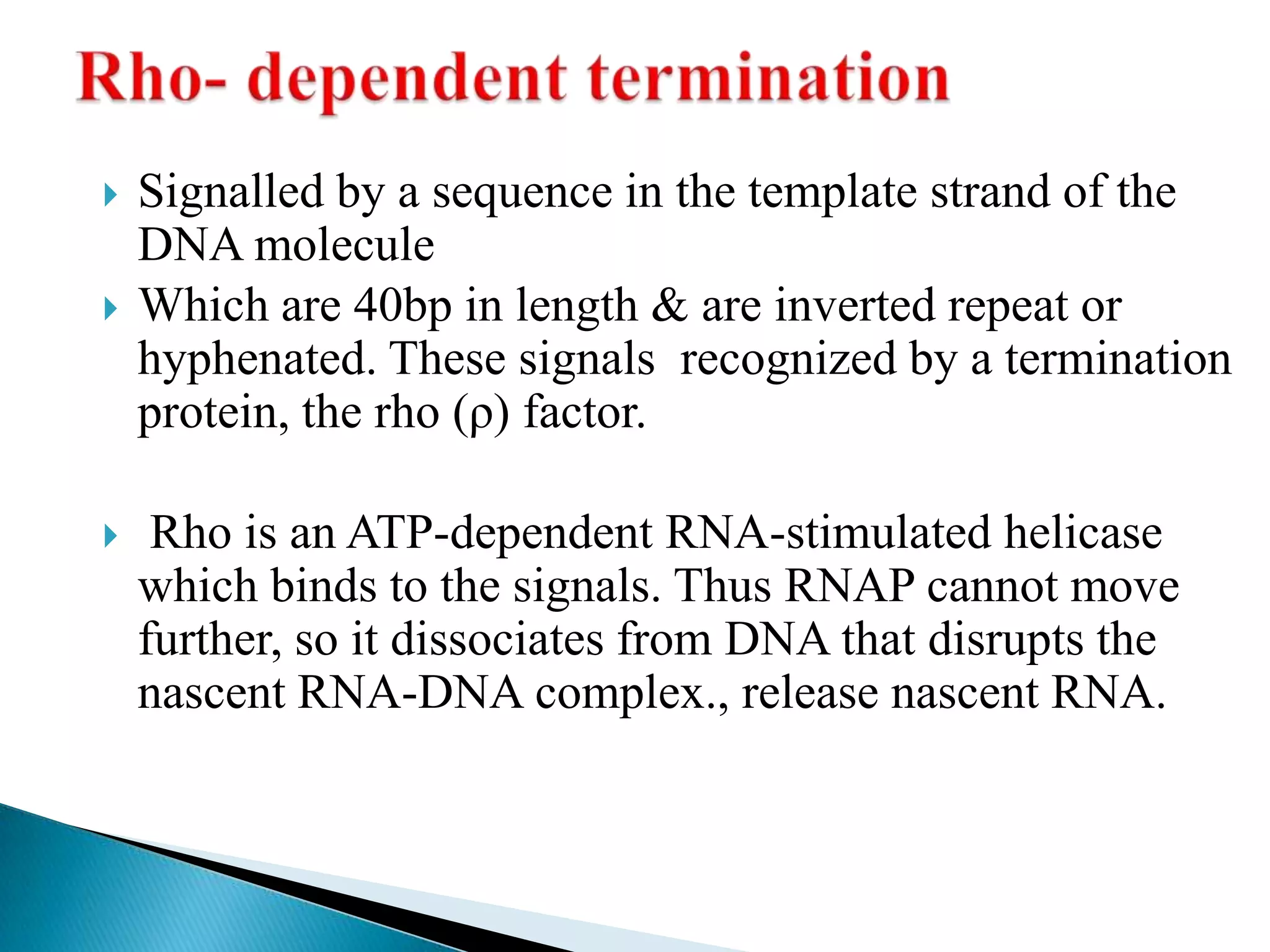
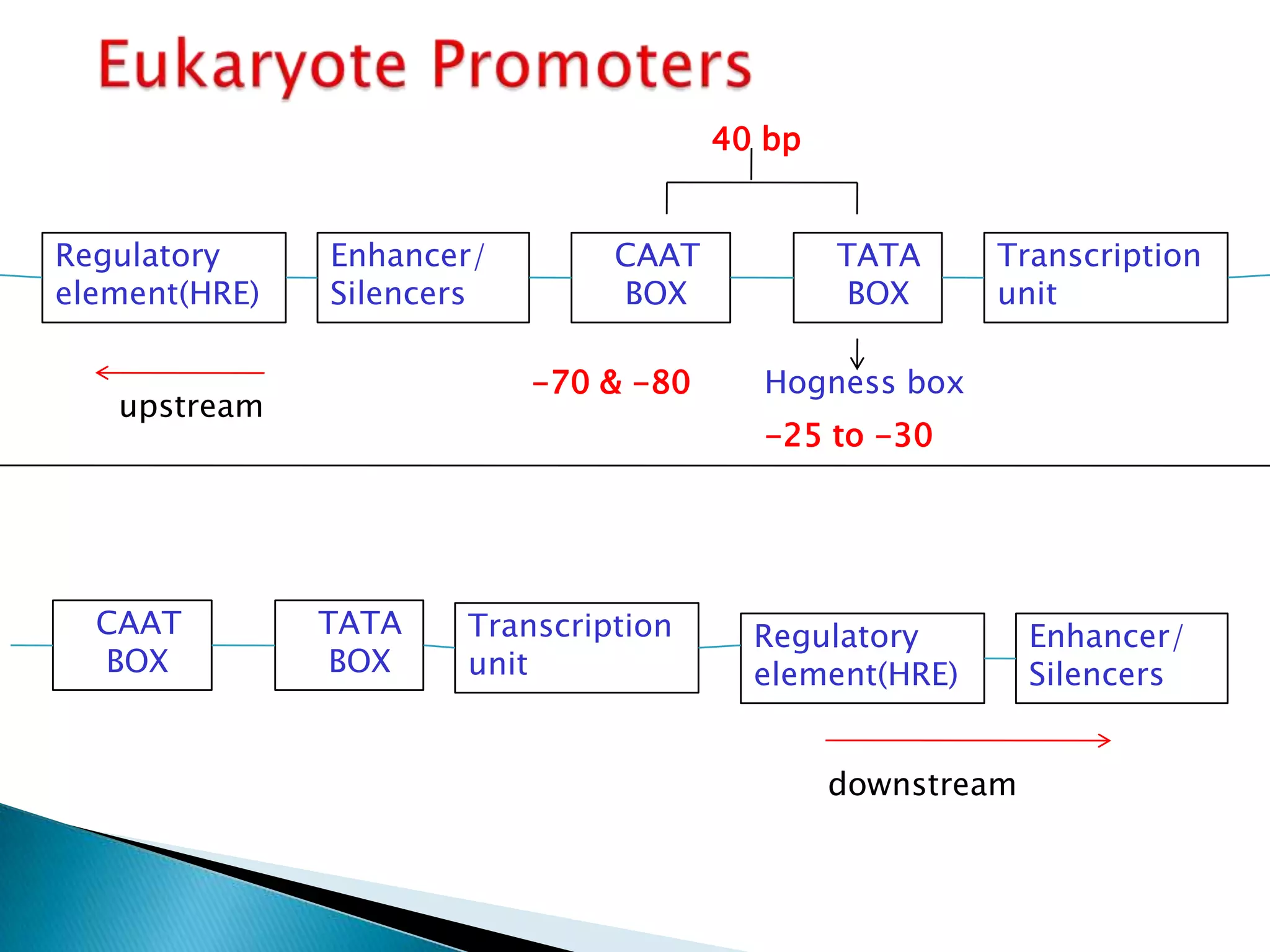
3. Transcription involves initiation, elongation, and termination stages. Initiation requires promoters, elongation uses RNA polymerase to add nucleotides, and termination ends RNA synthesis.