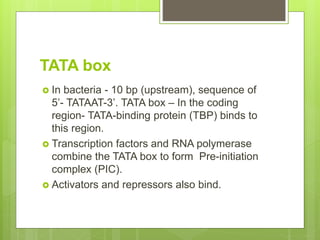
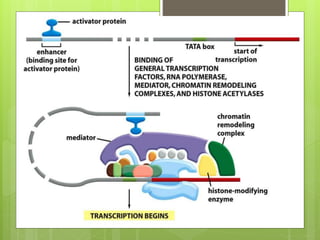
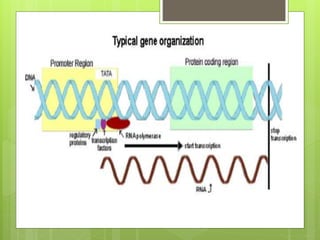
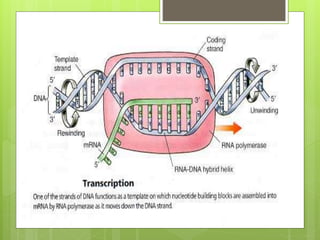
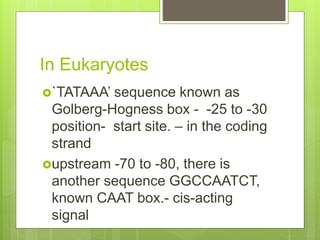
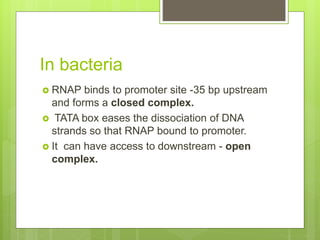
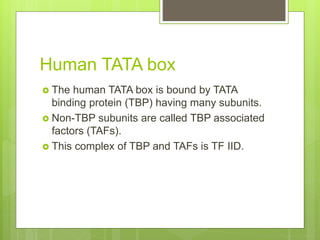
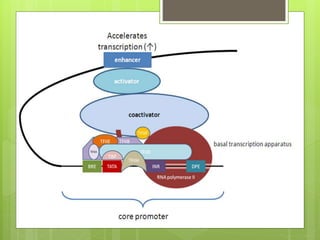
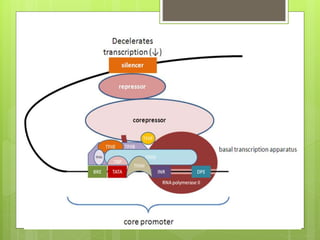
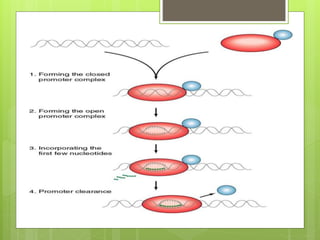
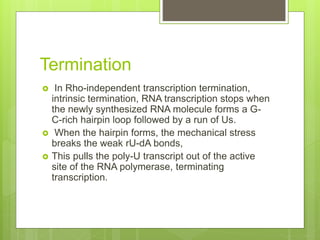
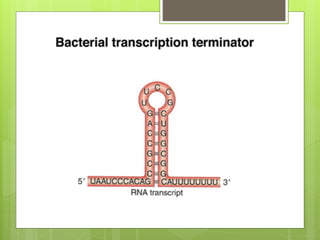
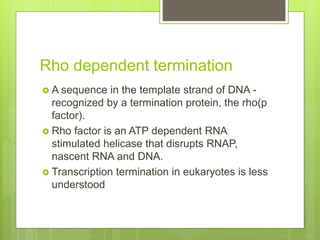
1. The document discusses RNA synthesis and processing, including the different types of RNA (mRNA, rRNA, tRNA), the process of transcription, initiation, elongation, and termination.
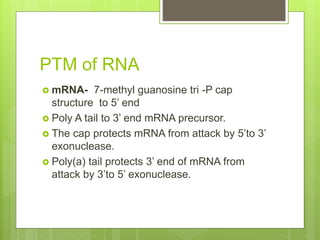
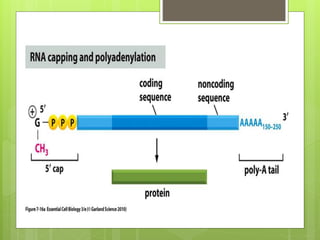
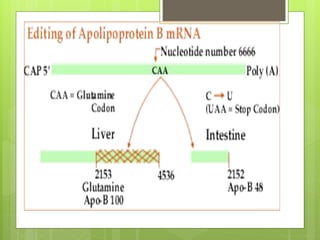
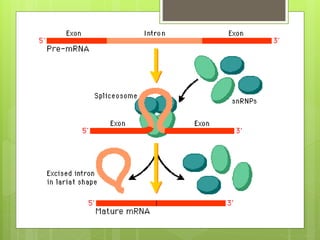
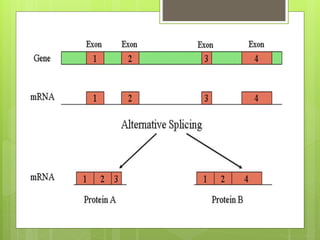
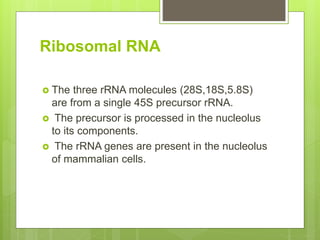
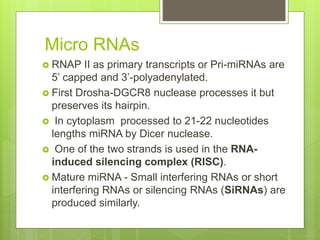
2. It also covers RNA processing after transcription, including 5' capping, polyadenylation, splicing, and modifications to tRNA, rRNA and other non-coding RNAs.
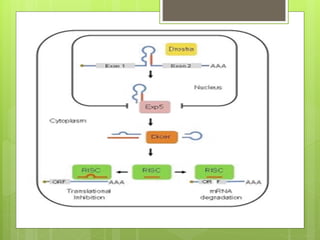
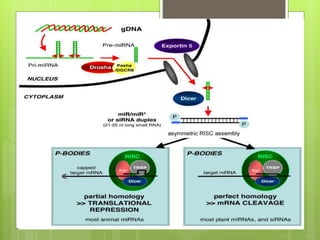
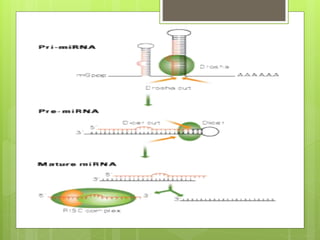
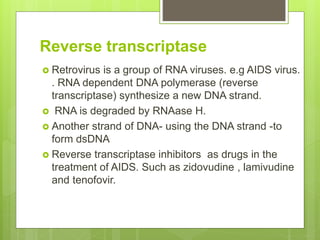
3. The clinical applications of understanding RNA synthesis and processing are discussed, such as targets for antibiotics, implications for genetic diseases, and miRNA roles in various human health conditions.