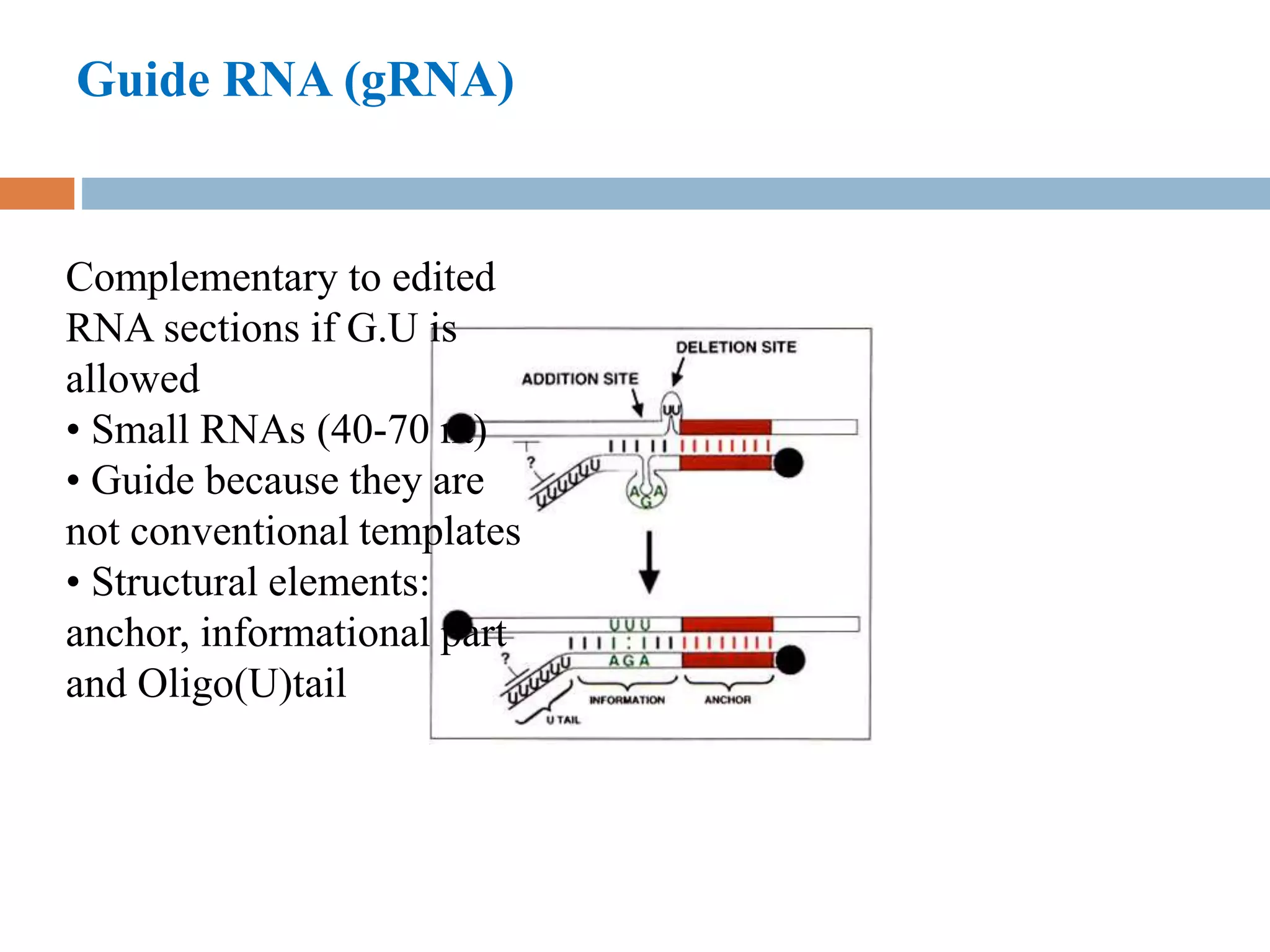
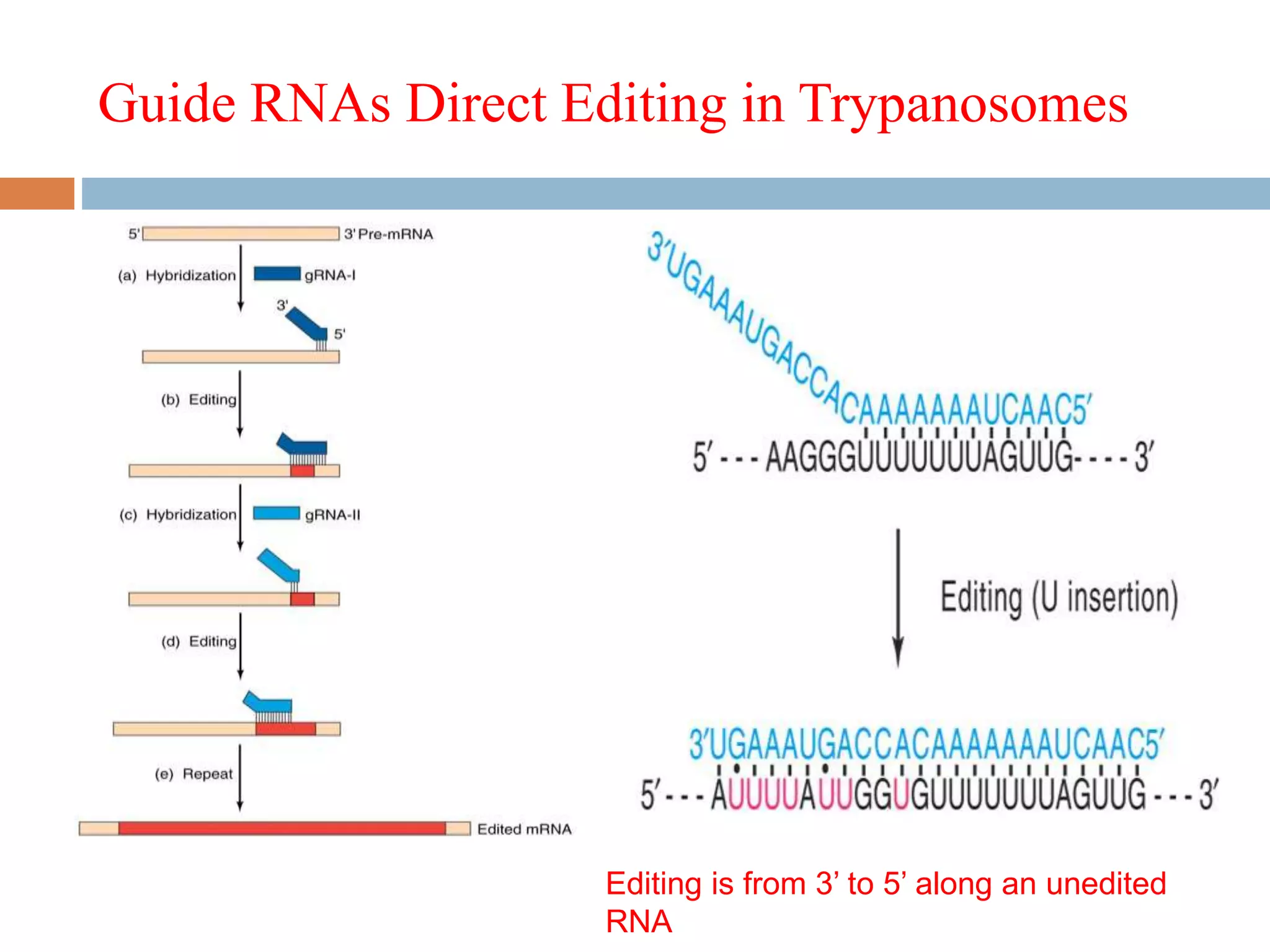
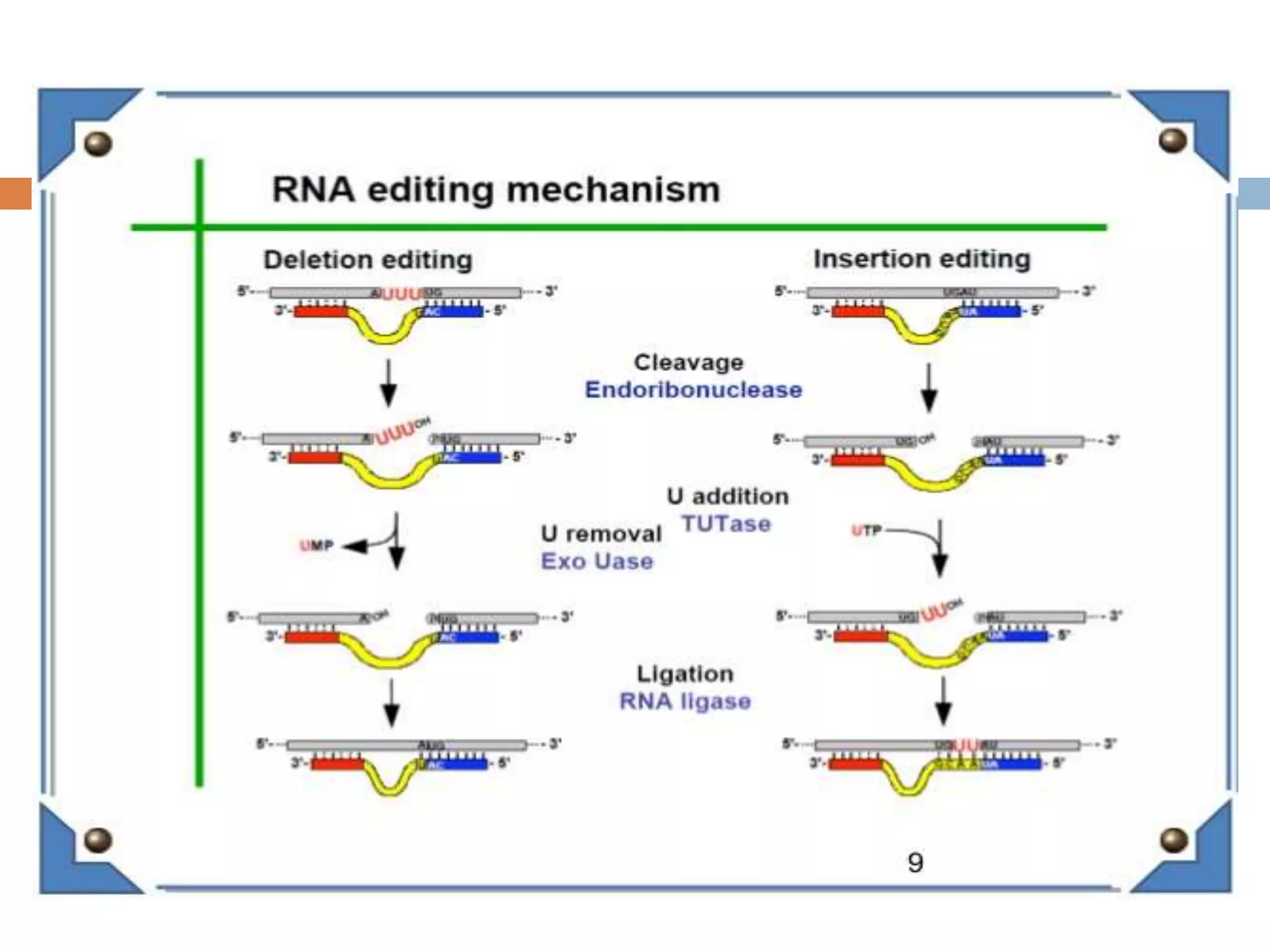
RNA editing is a process that alters RNA sequences to differ from DNA templates, primarily observed in protozoa, plants, and mammals. In trypanosomes, specifically, pre-mRNAs undergo site-specific insertions and deletions of uridylate residues, allowing for the creation of functional proteins from pseudogenes. This editing involves small guide RNAs that direct the editing machinery, catalyzing changes from the 3' to 5' direction and incorporating various enzymatic activities.