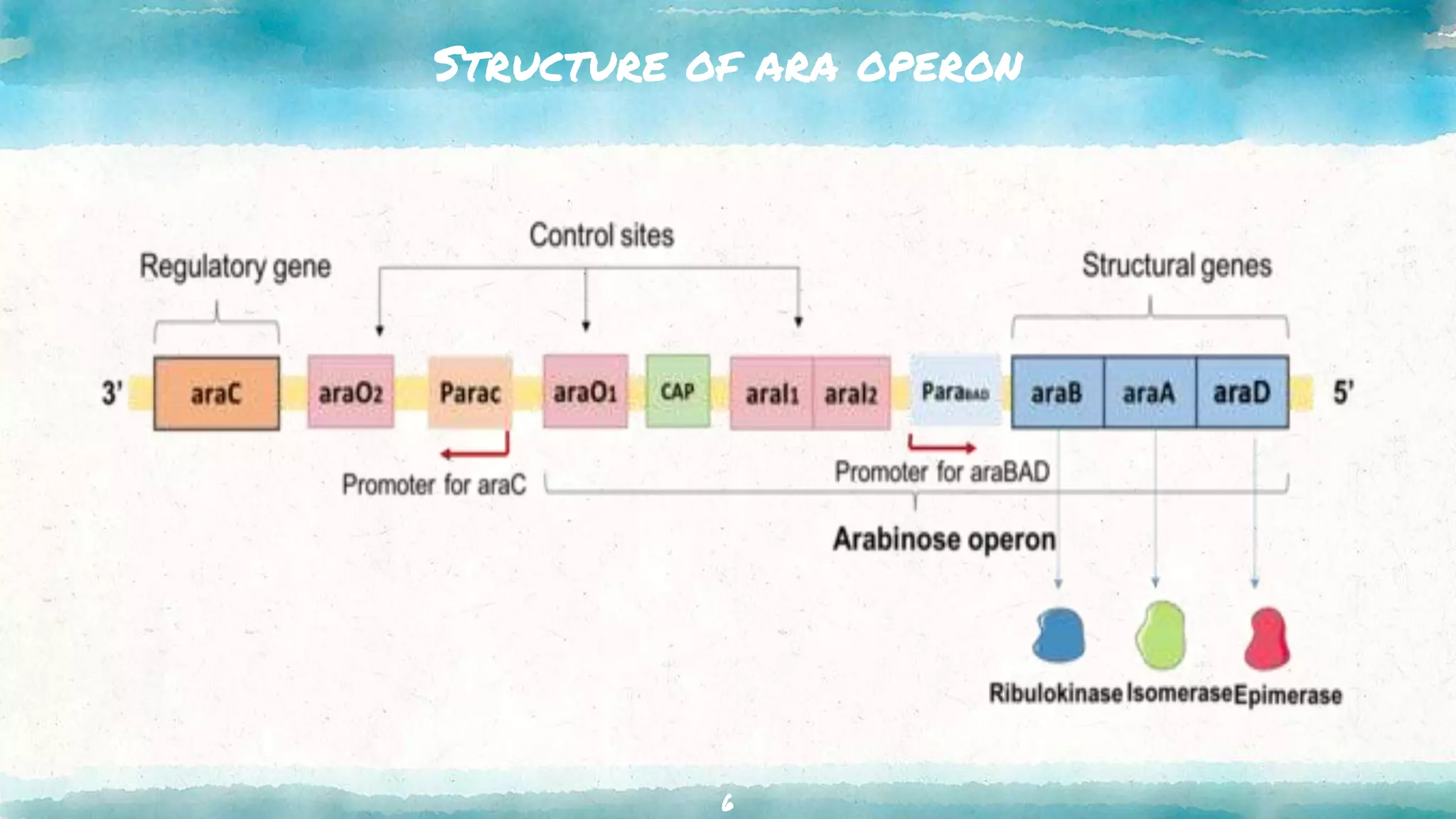
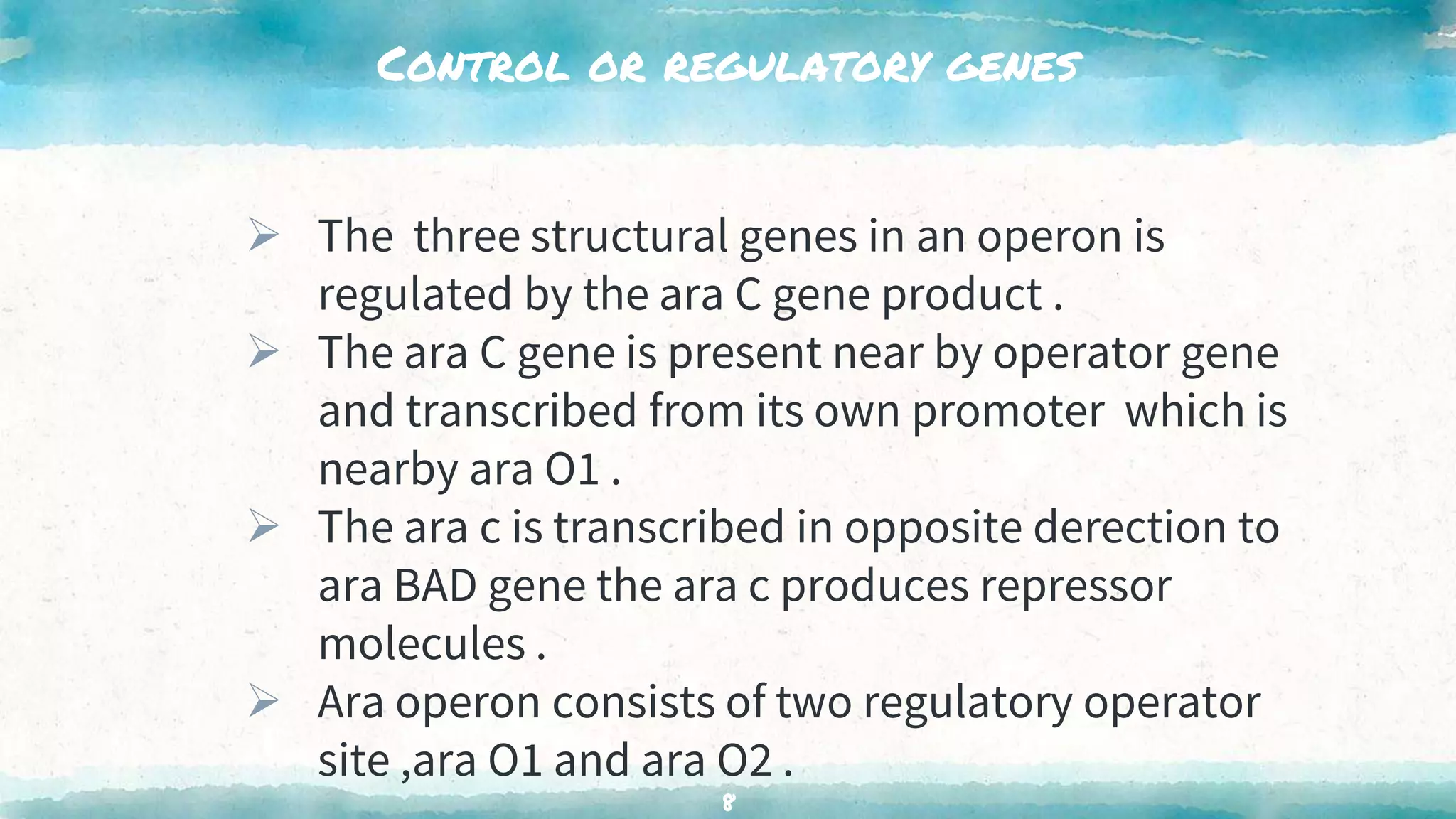
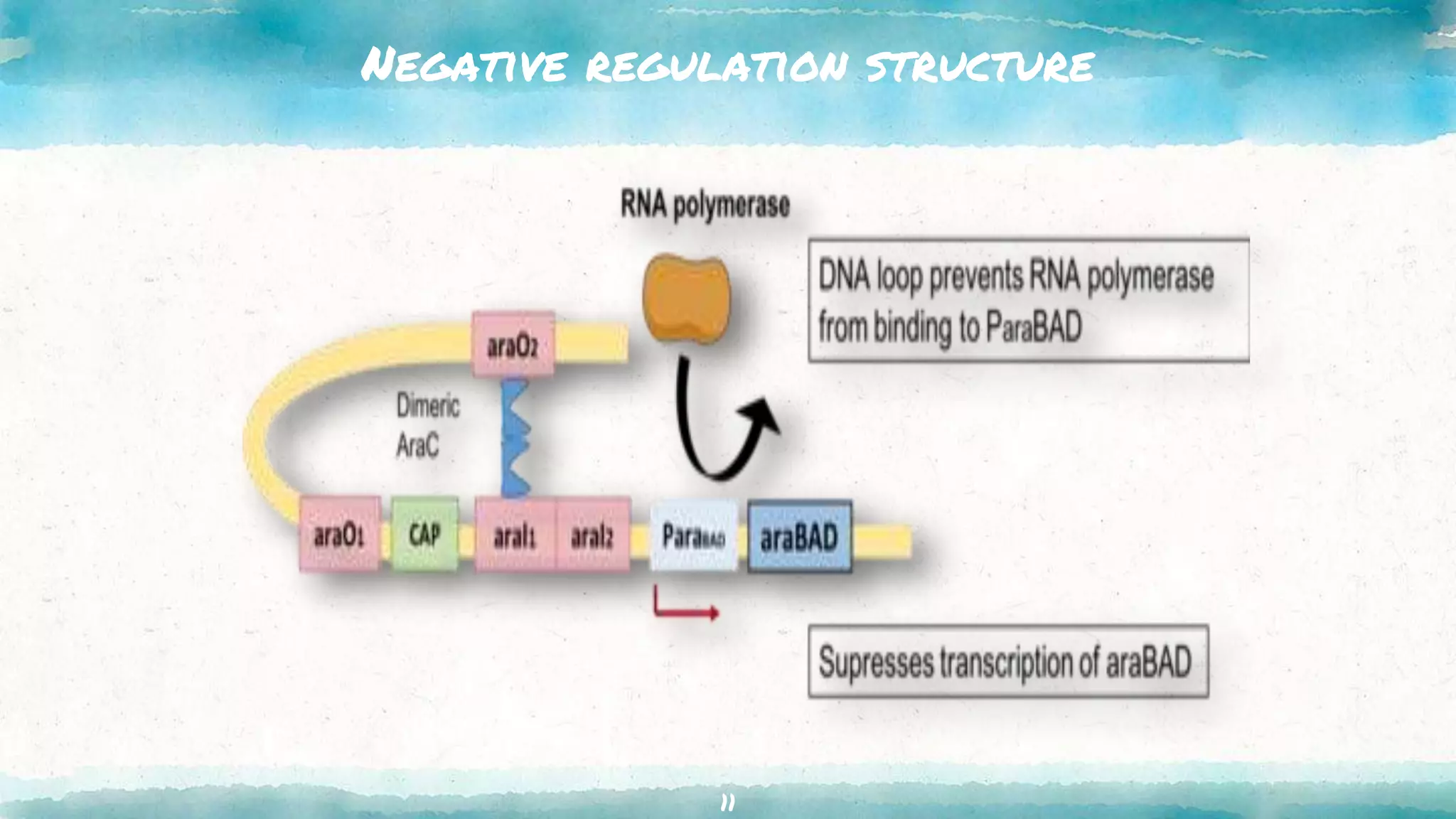
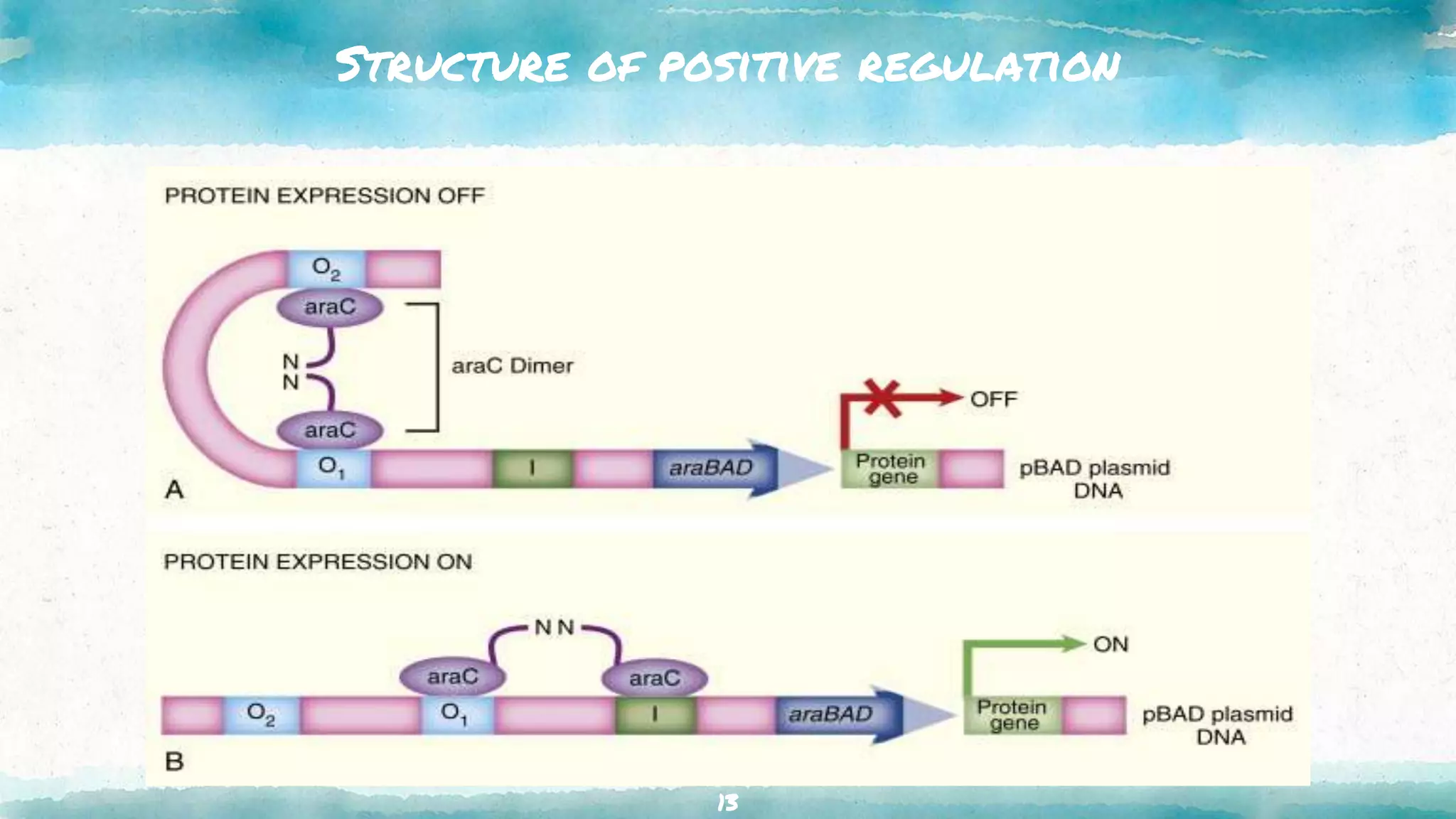
The document summarizes the ara operon, which regulates genes involved in the metabolism of arabinose sugar. It consists of 3 key components: structural genes that encode enzymes for arabinose catabolism, an operator site that binds a repressor protein, and a promoter that enables transcription. Specifically, the ara operon contains 3 structural genes - araB, araA, araD - that code for enzymes converting arabinose into an intermediate. It is regulated by the araC gene product, which can act as both a repressor and activator depending on arabinose and glucose levels. When arabinose is low and glucose high, the araC repressor binds the operator sites, forming a DNA loop that