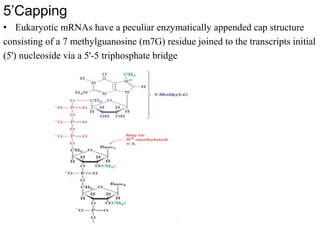
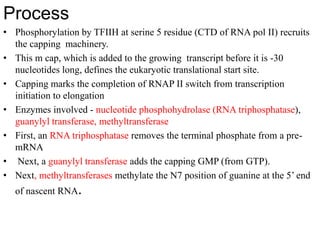
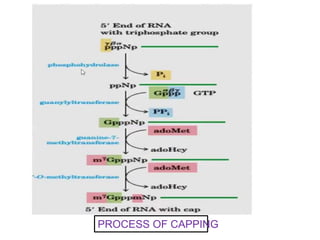
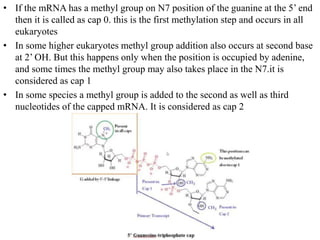
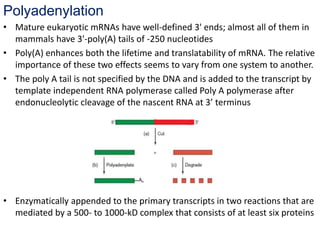
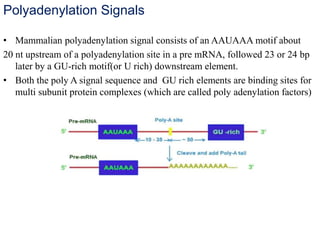
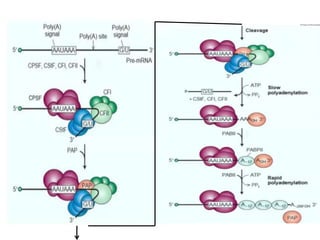
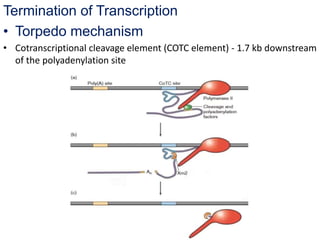
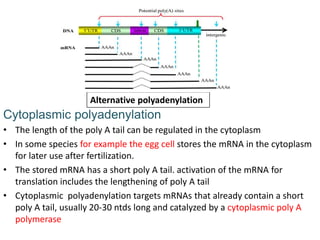
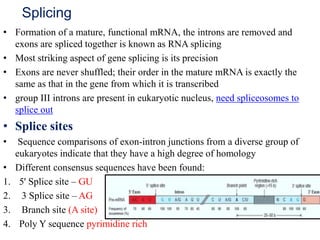
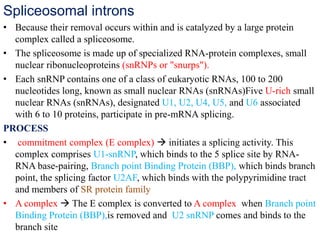
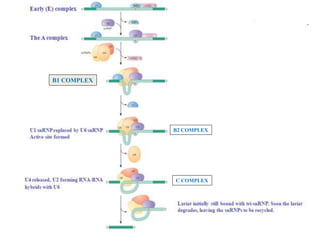
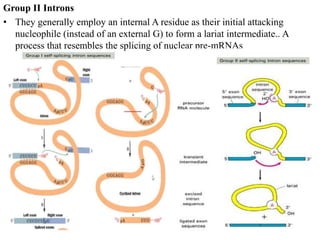
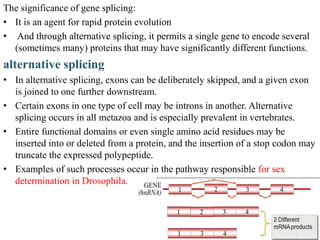
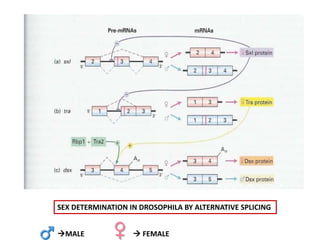
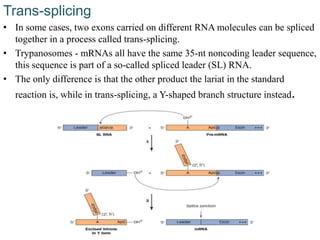
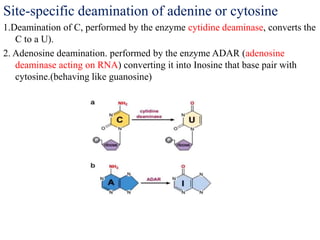
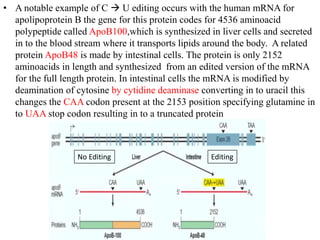
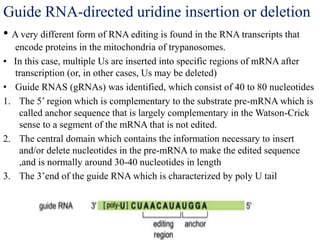
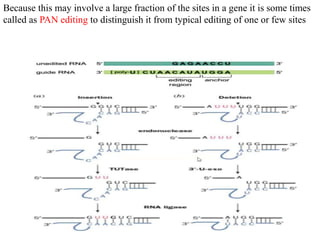
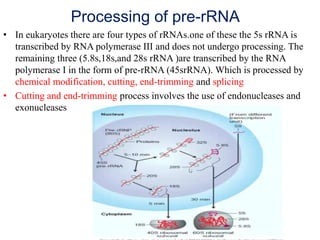
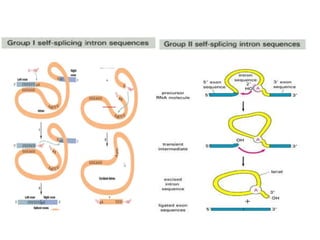
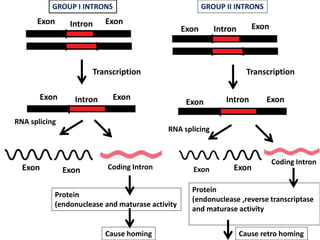
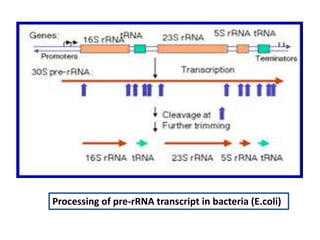
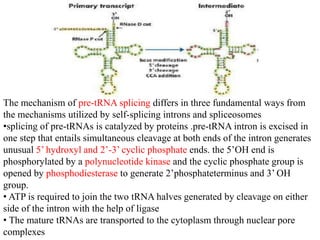
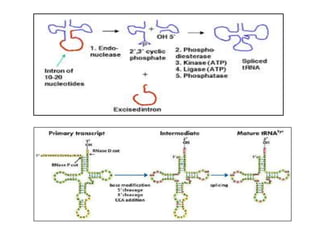
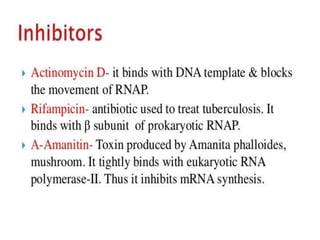
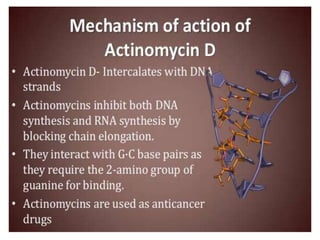
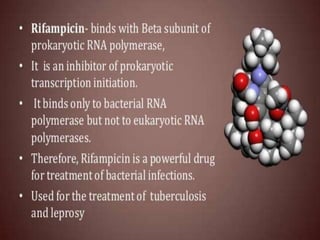
The document discusses RNA processing in eukaryotes versus prokaryotes, highlighting that eukaryotic pre-mRNA undergoes extensive modifications such as 5' capping, polyadenylation, and splicing, while prokaryotic mRNA is typically not modified. It details the steps and machinery involved in capping and polyadenylation, the intricacies of splicing including self-splicing introns, and the significance of these processes. Additionally, it outlines RNA editing mechanisms and the processing of pre-rRNA, summarizing the importance of these modifications in gene expression and protein diversity.