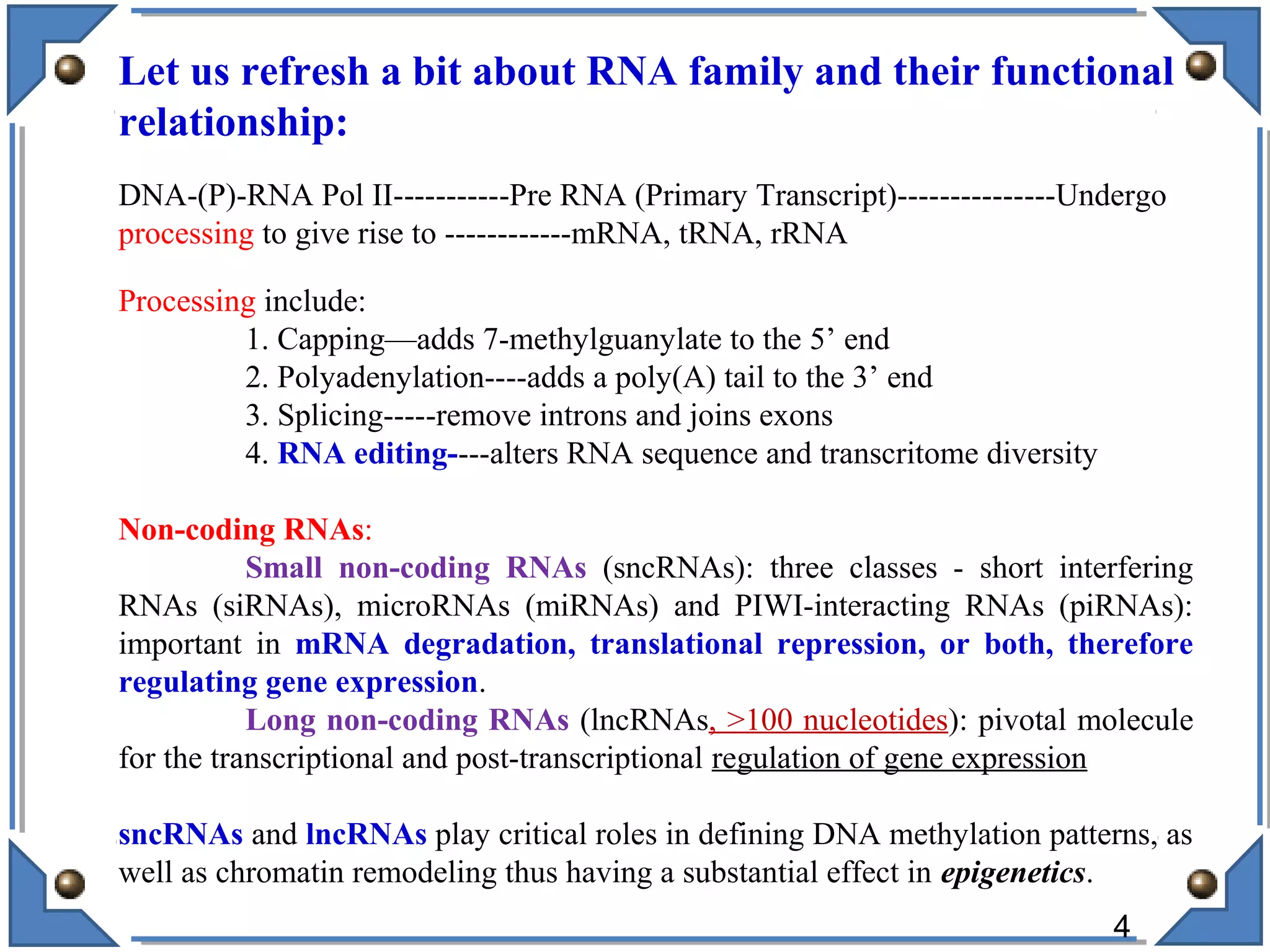
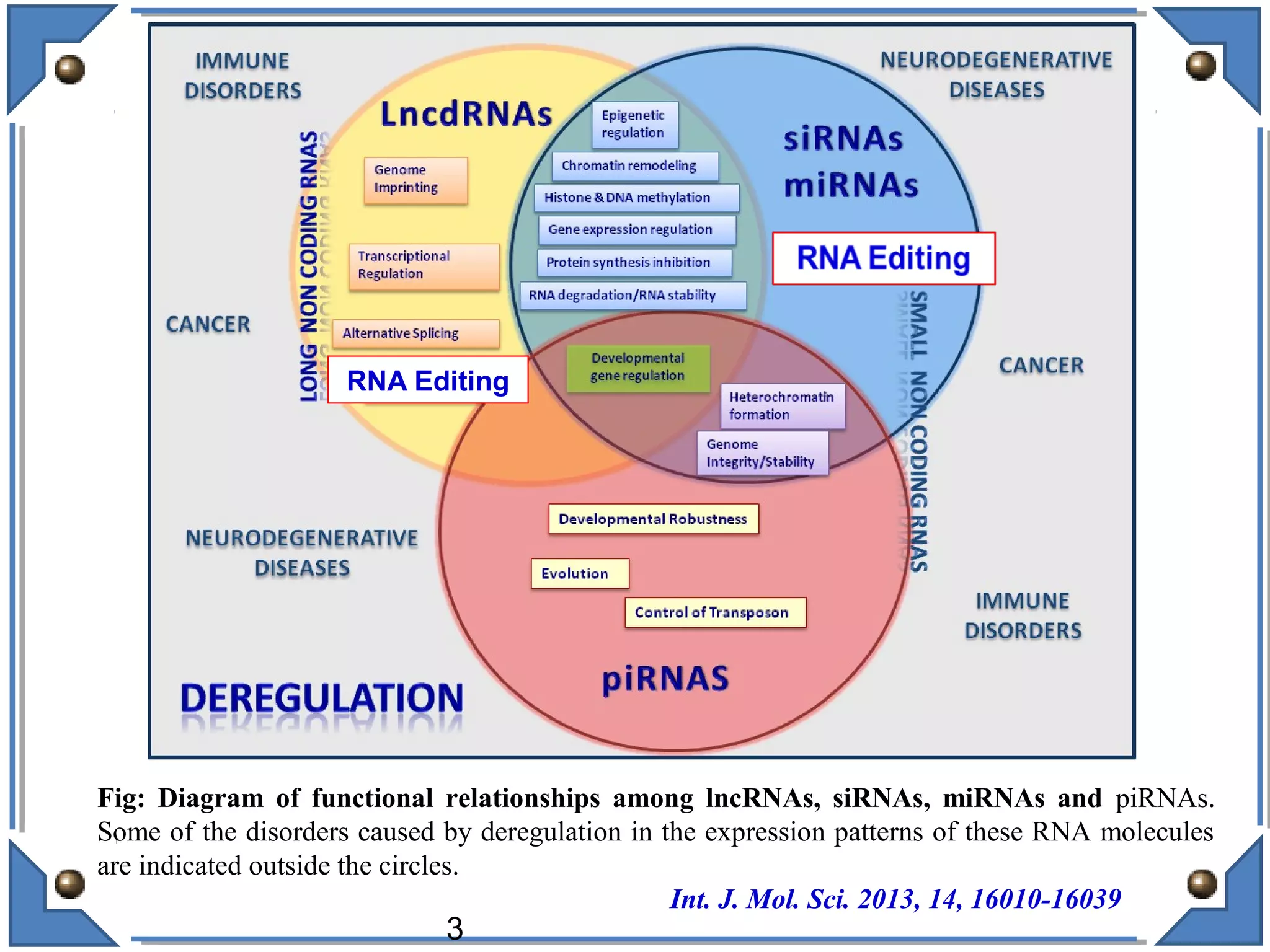
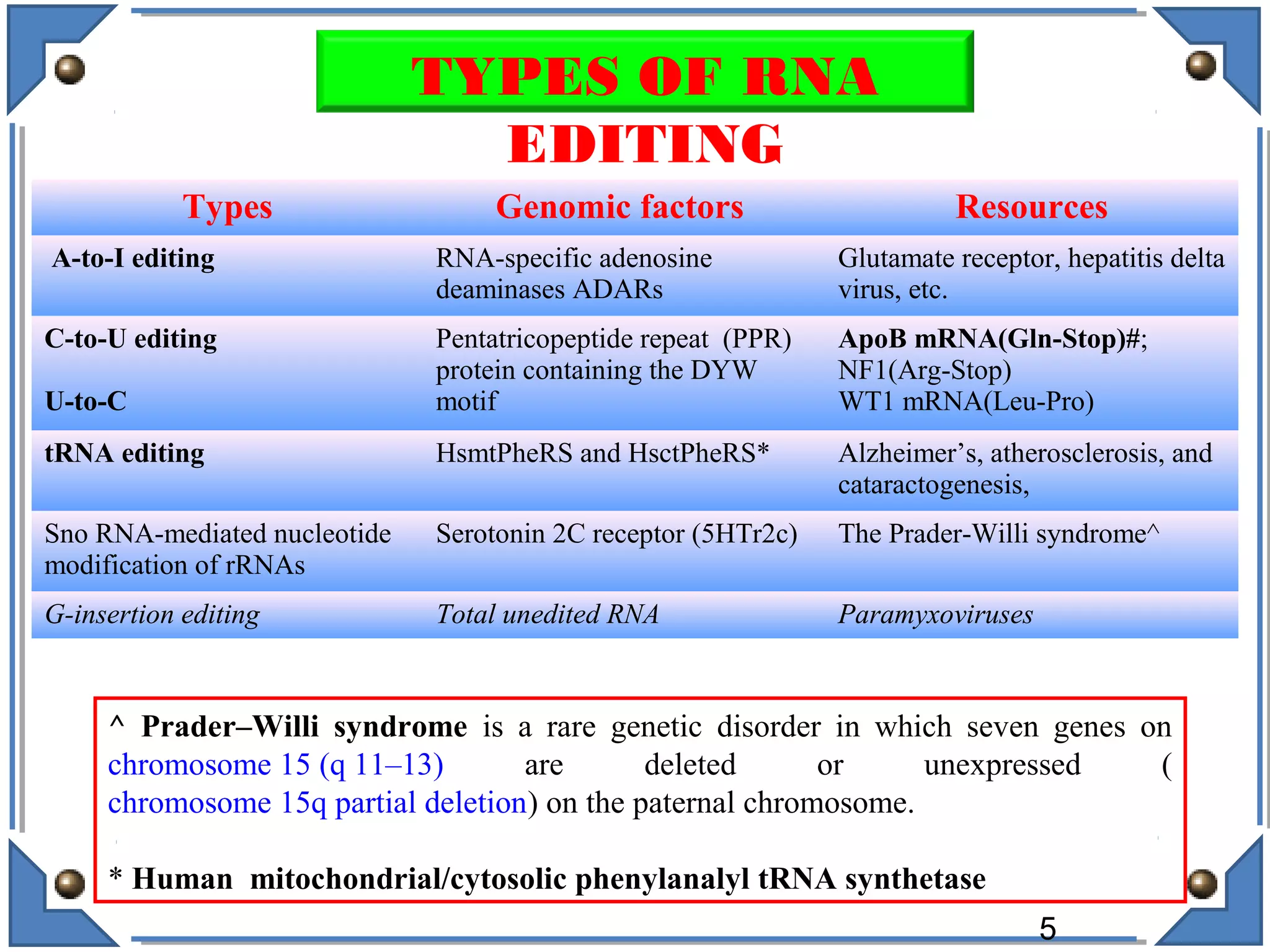
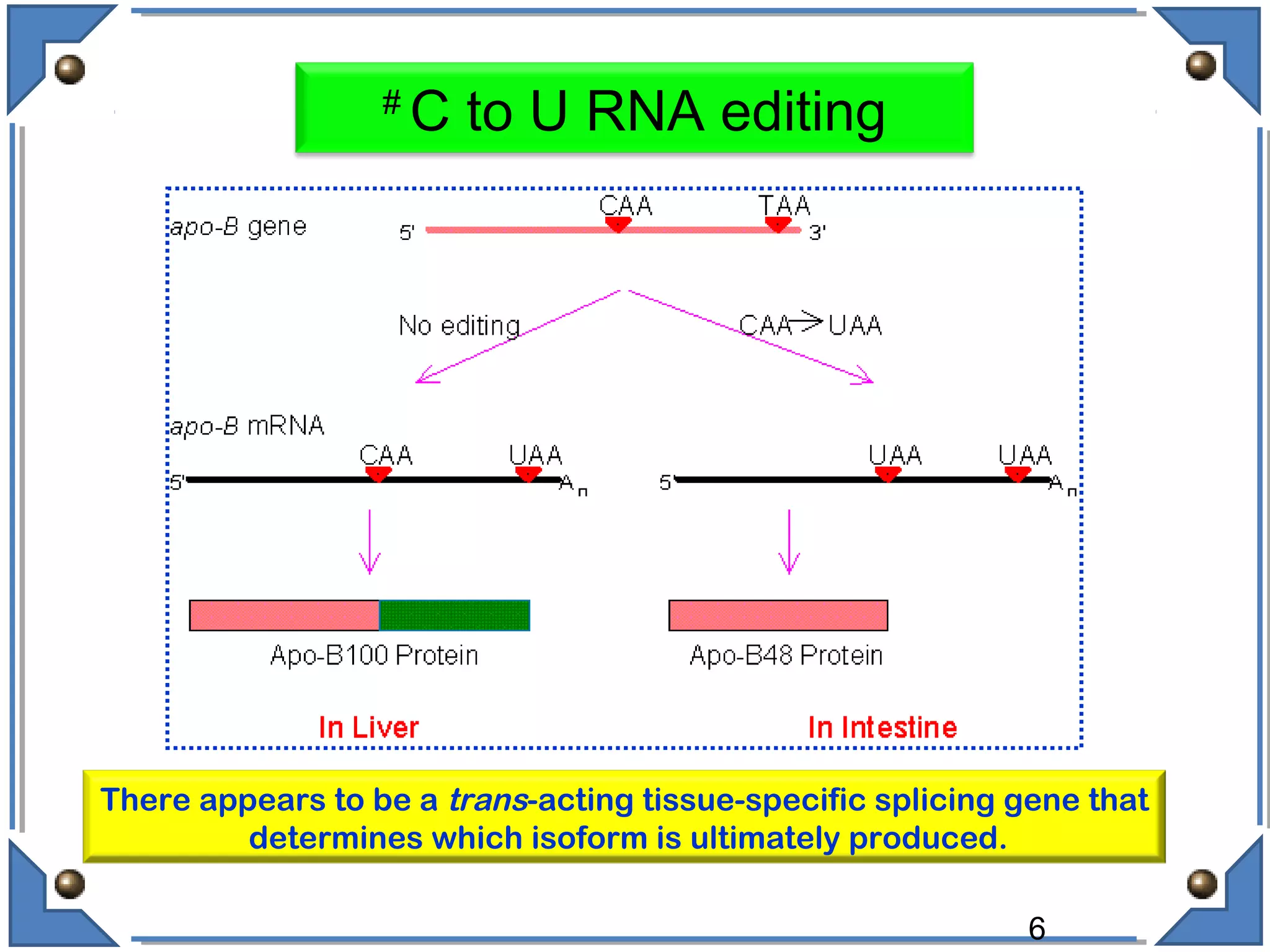
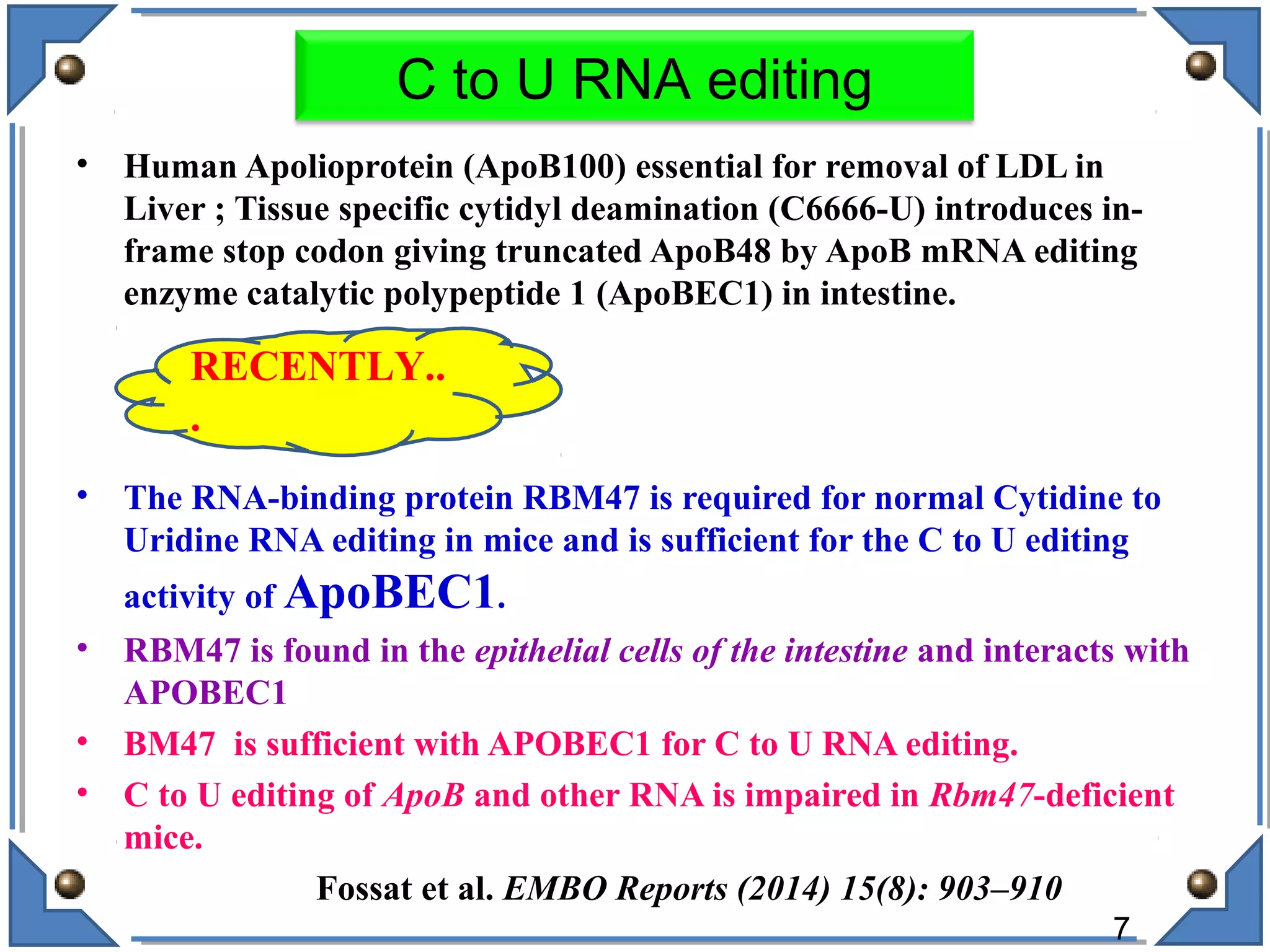
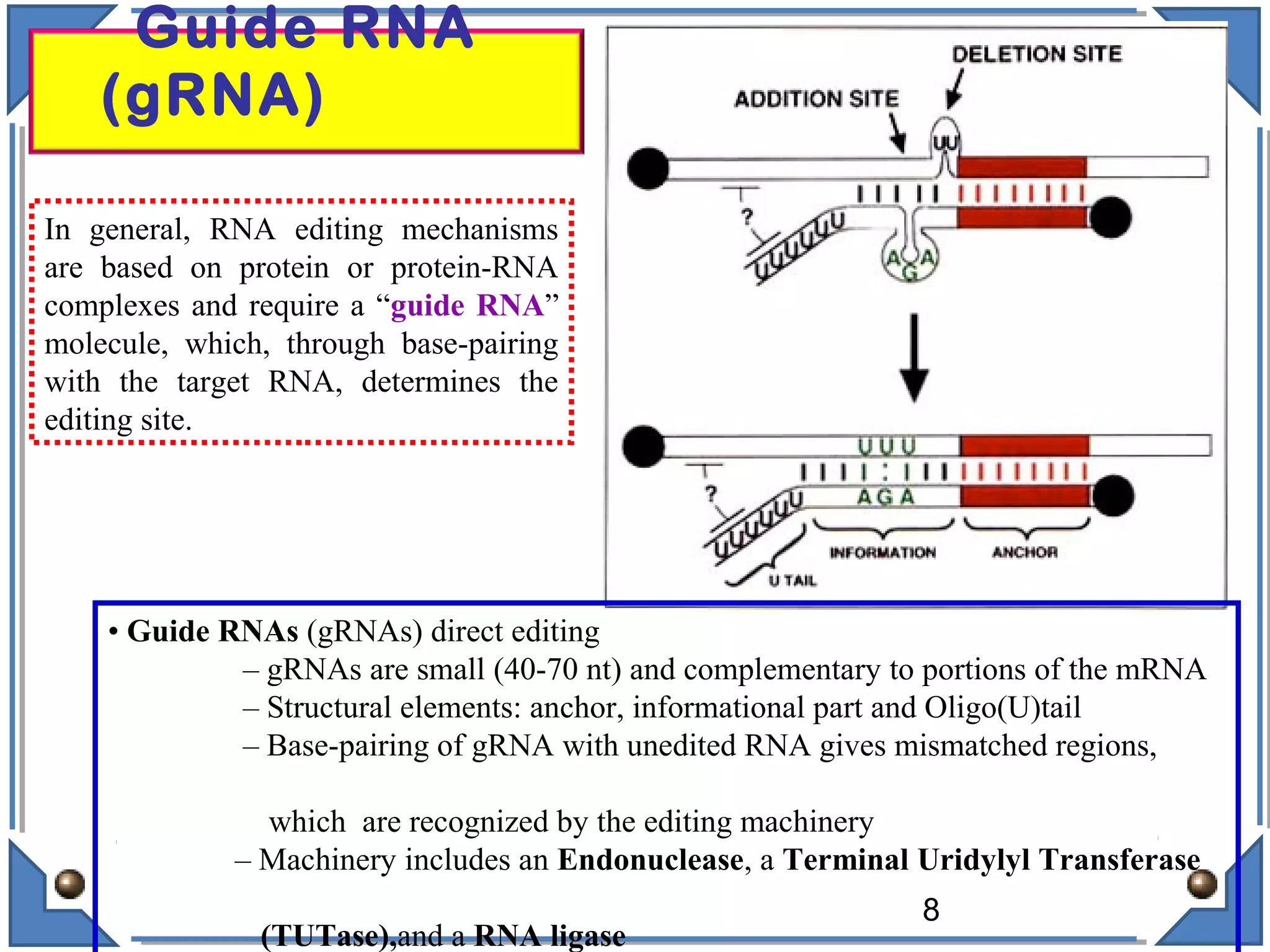
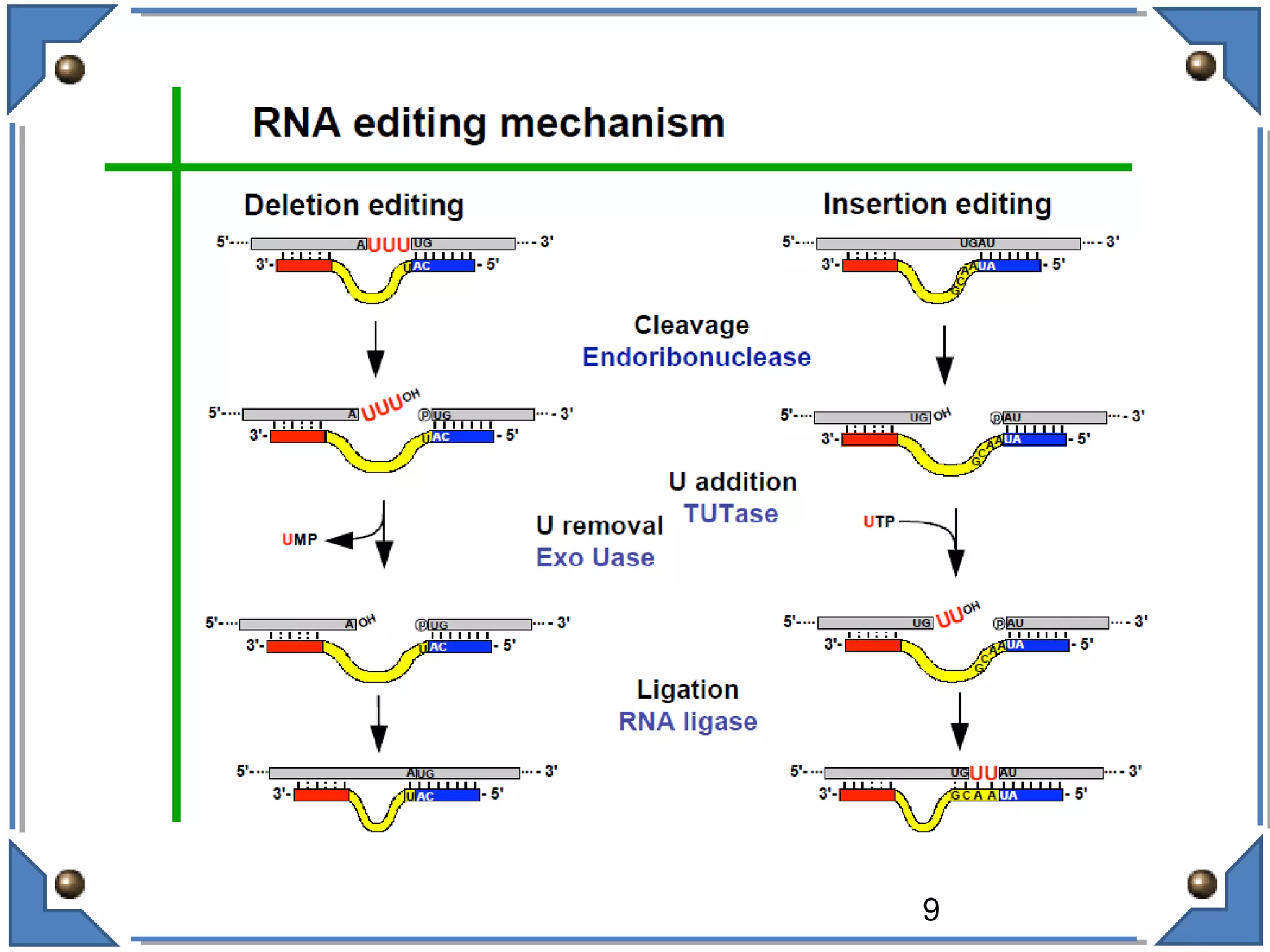
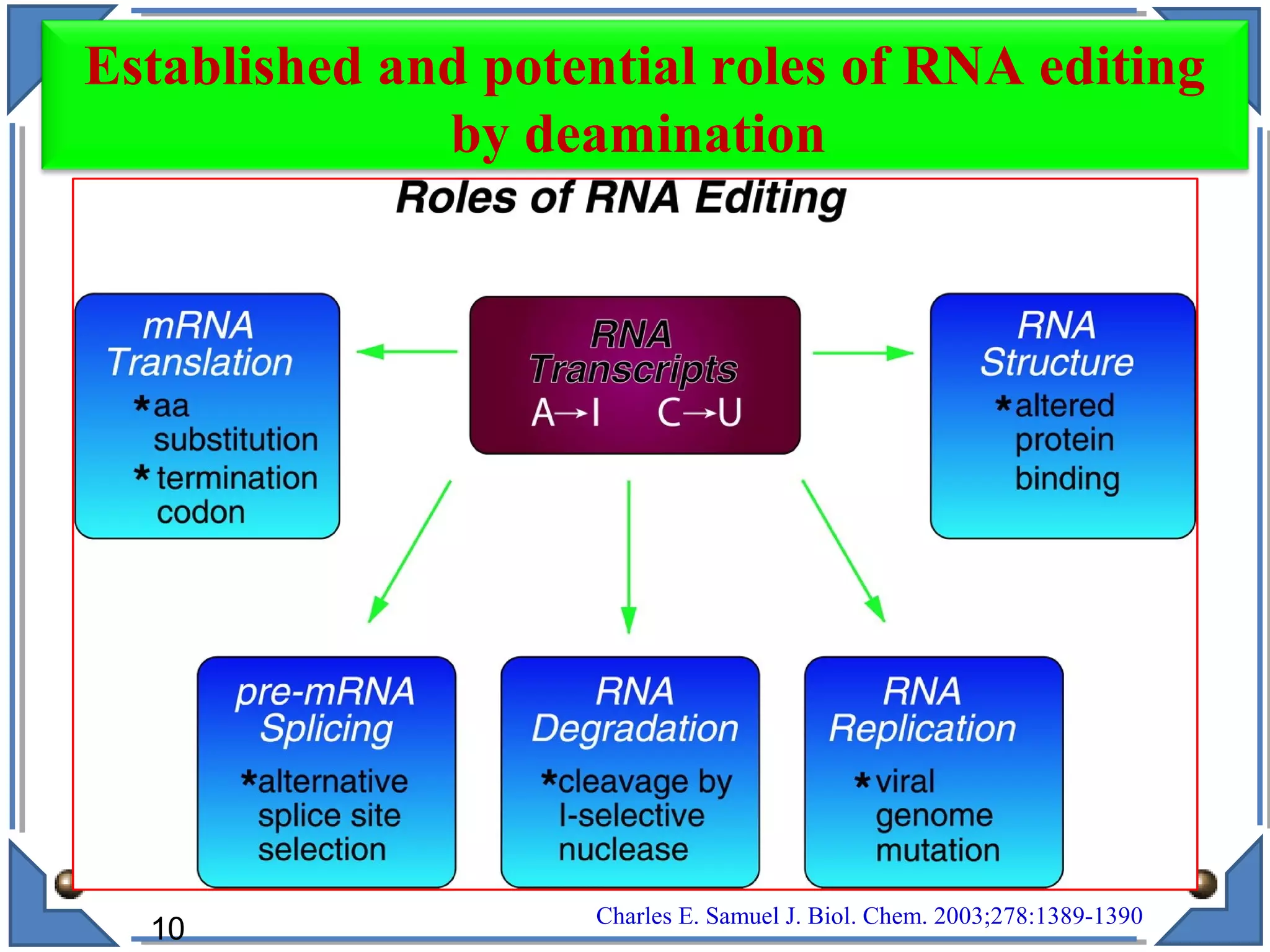
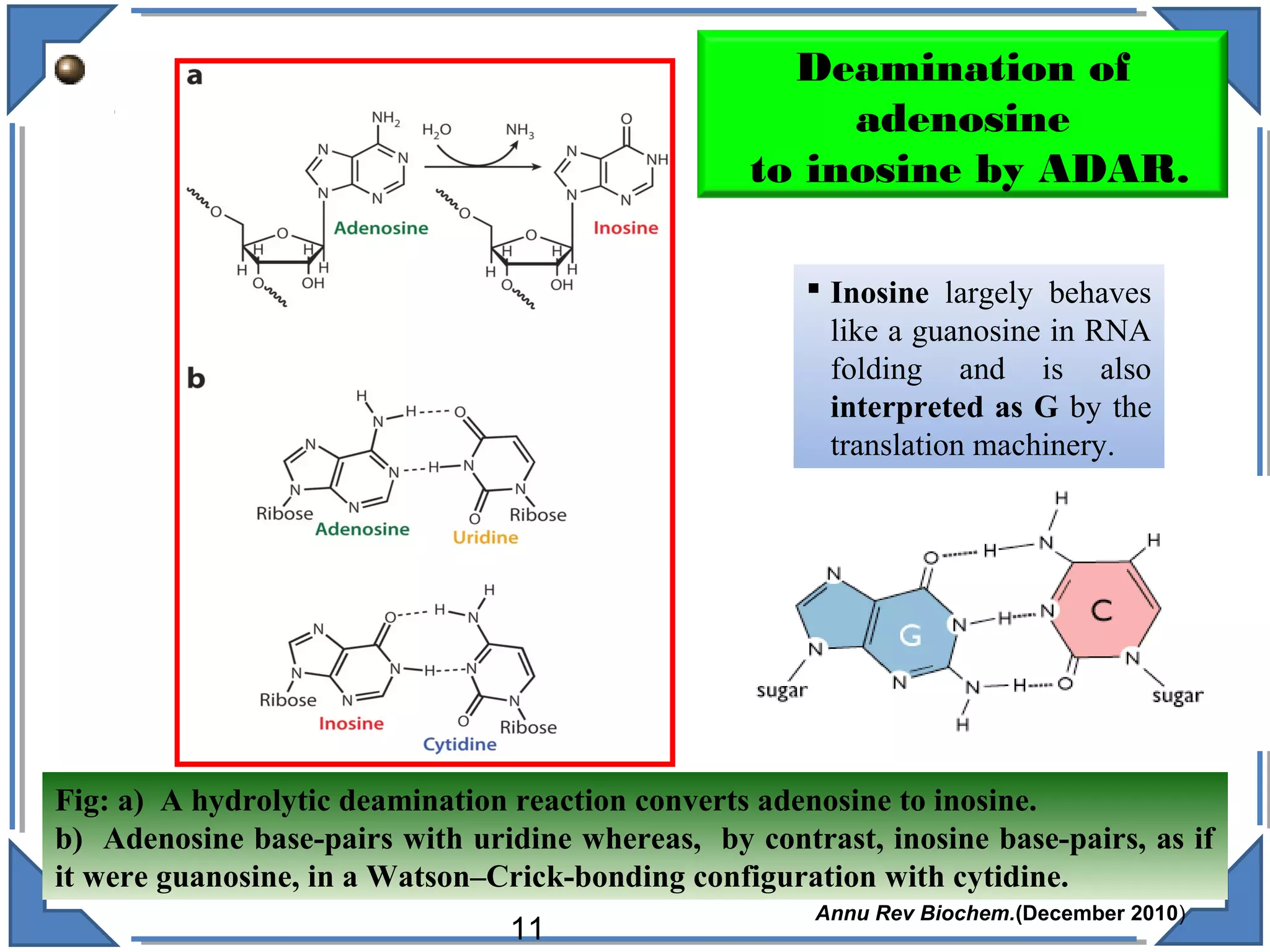
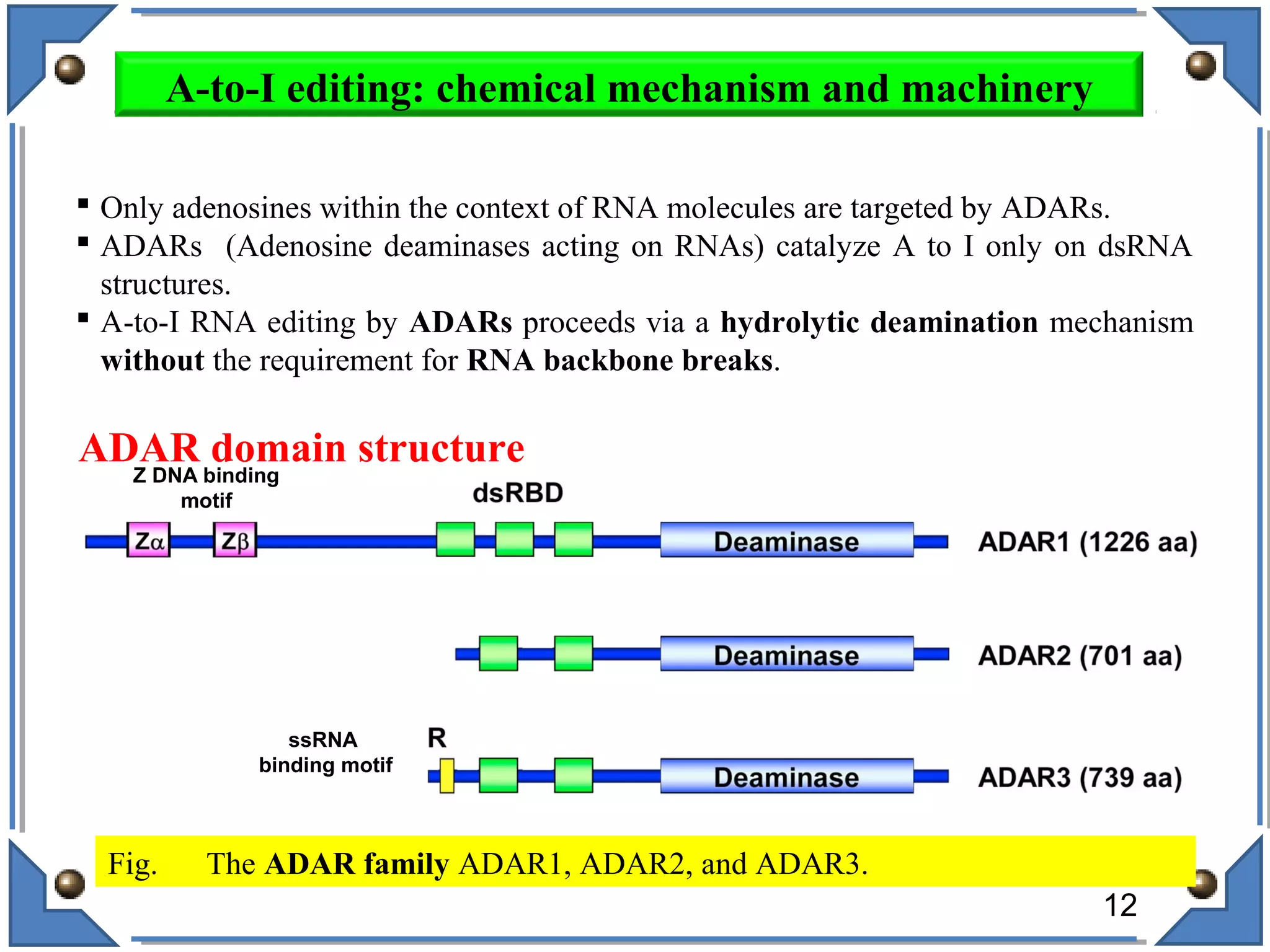
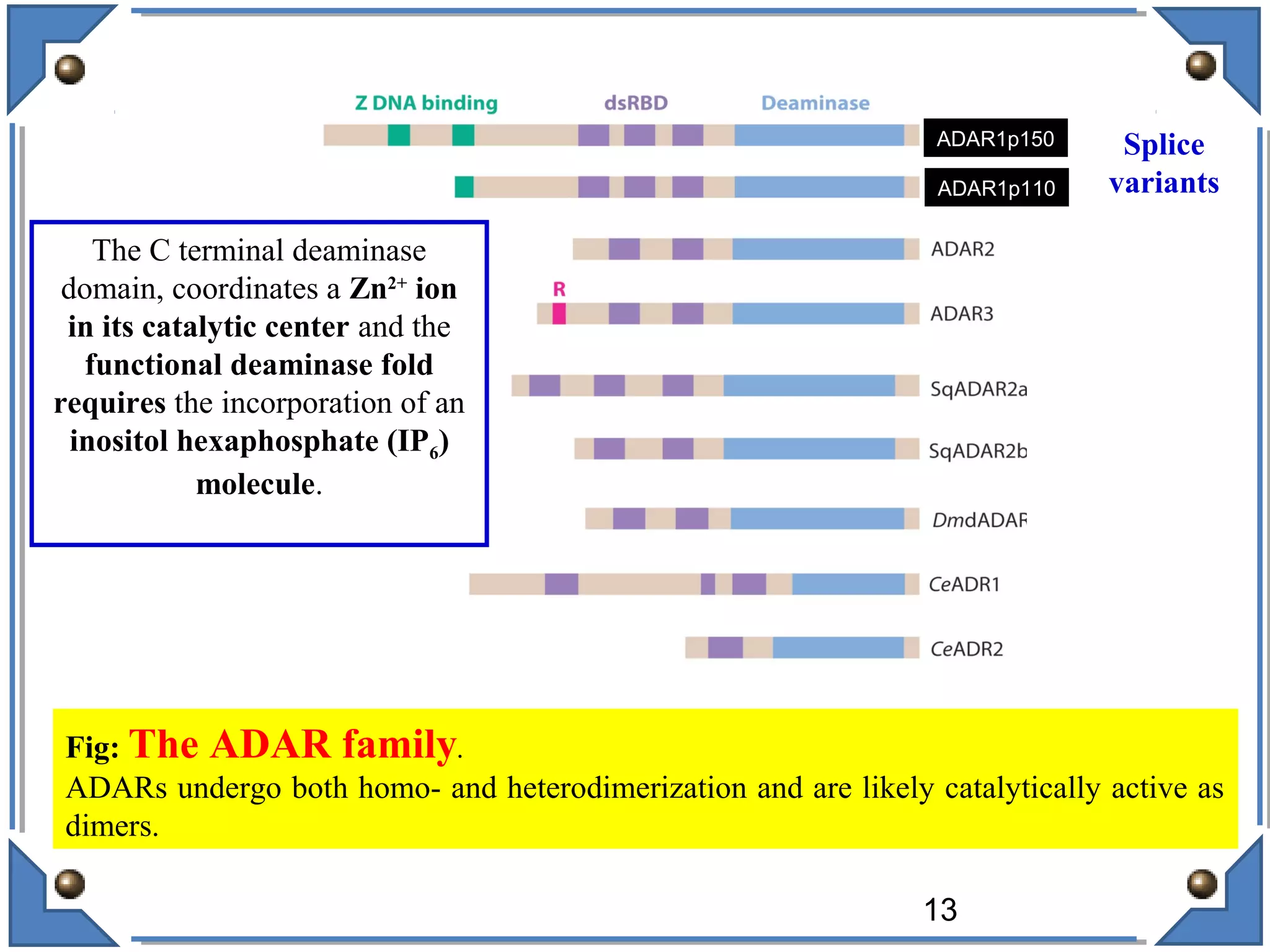
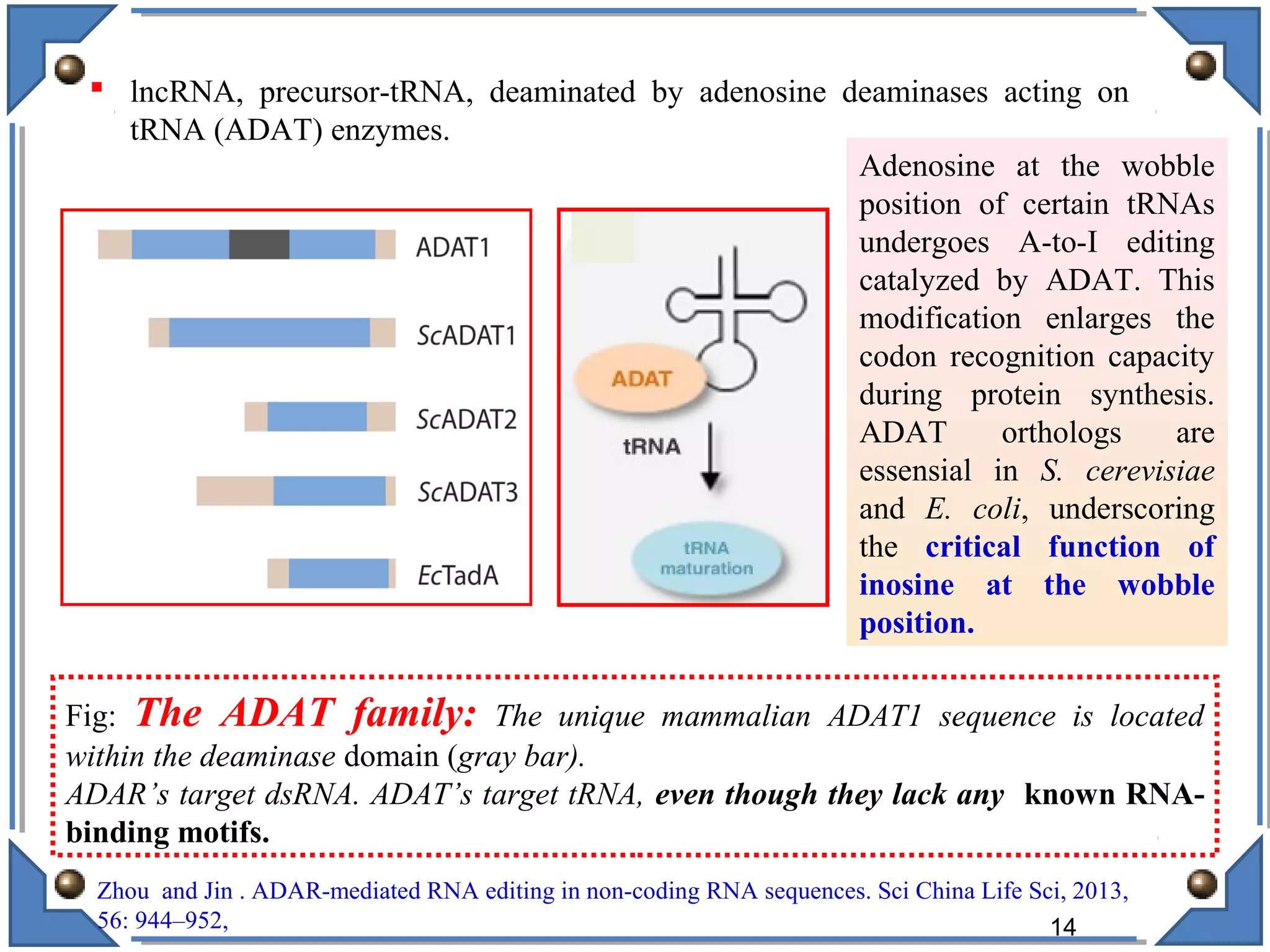
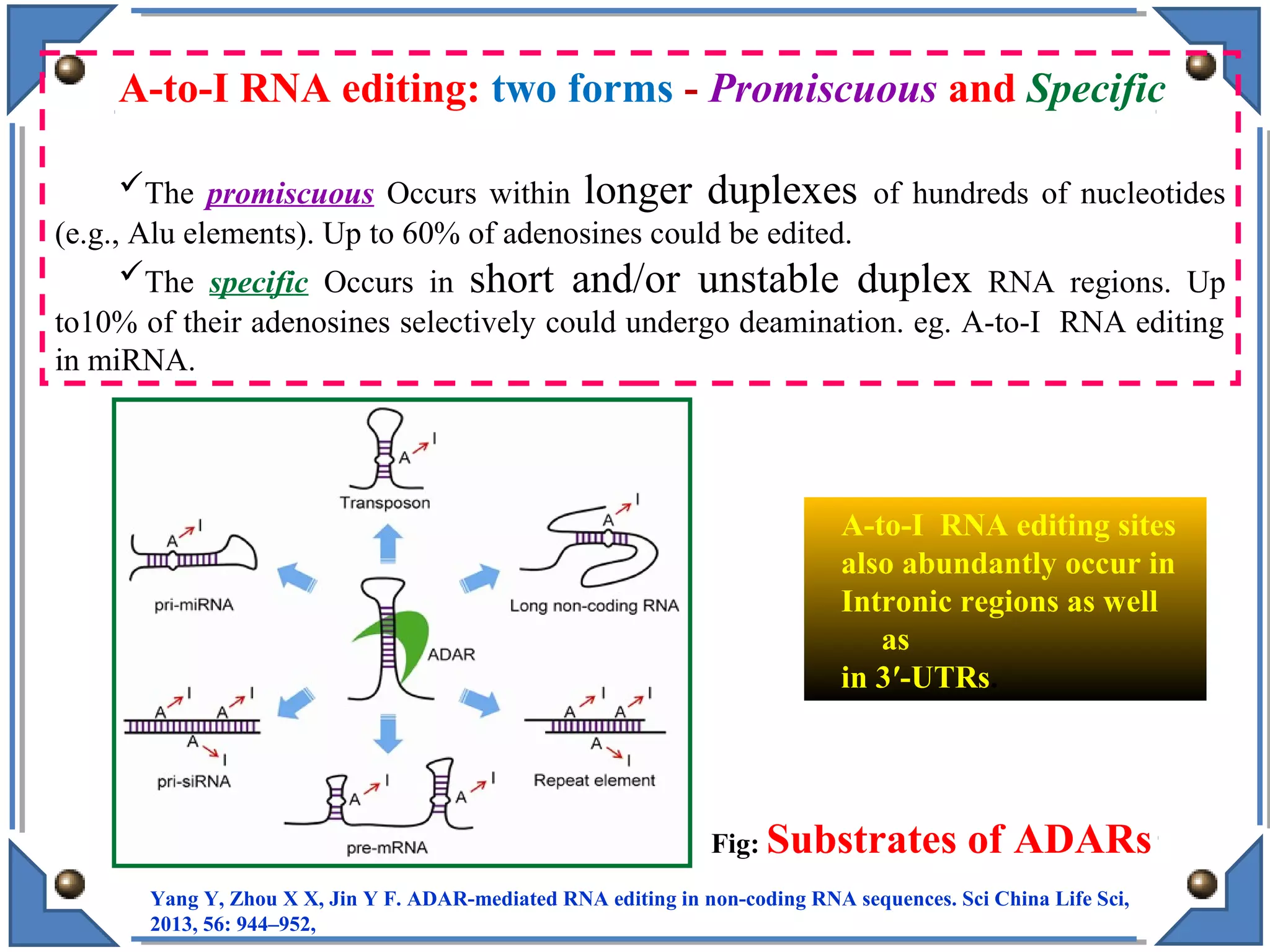
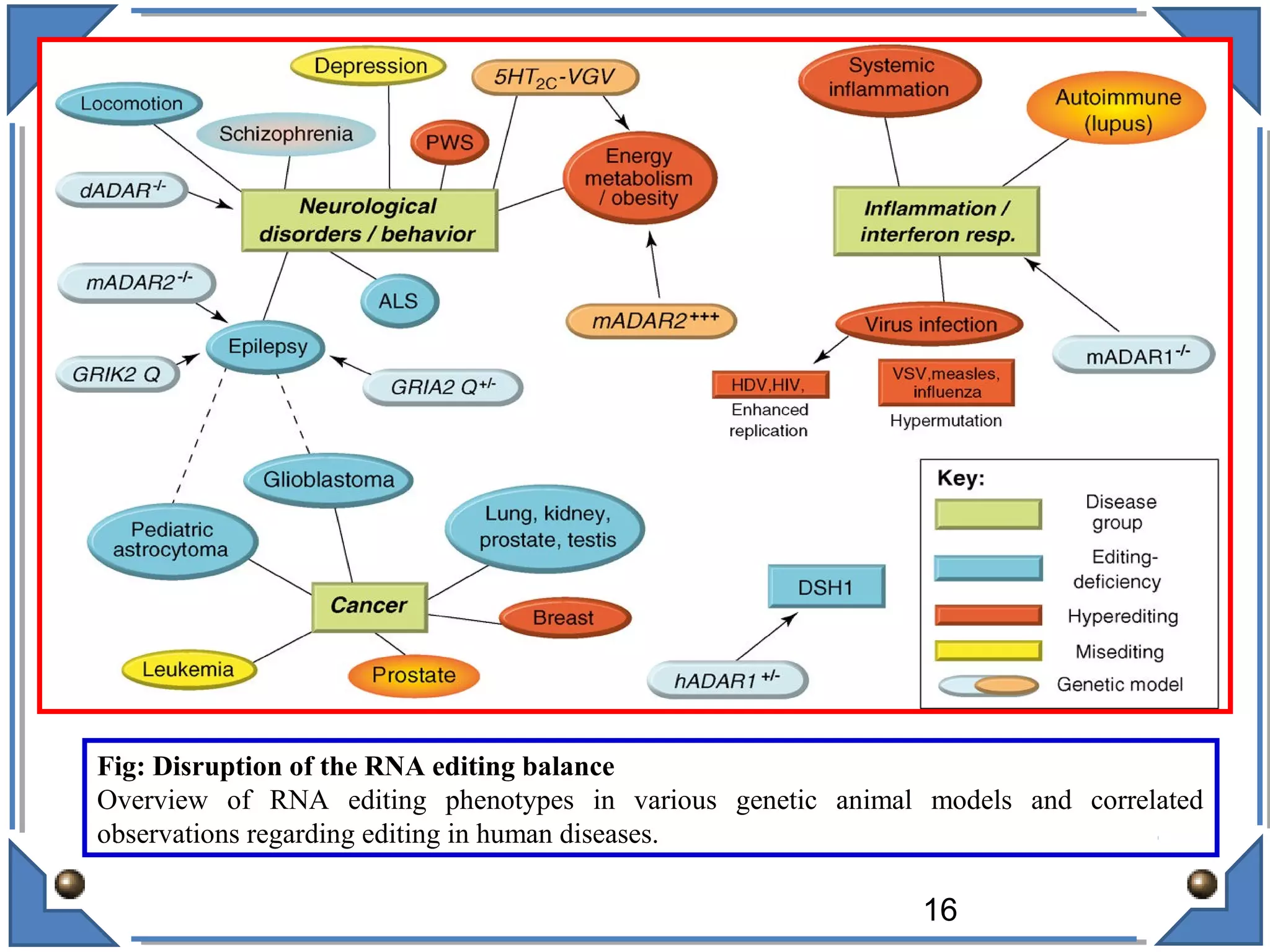
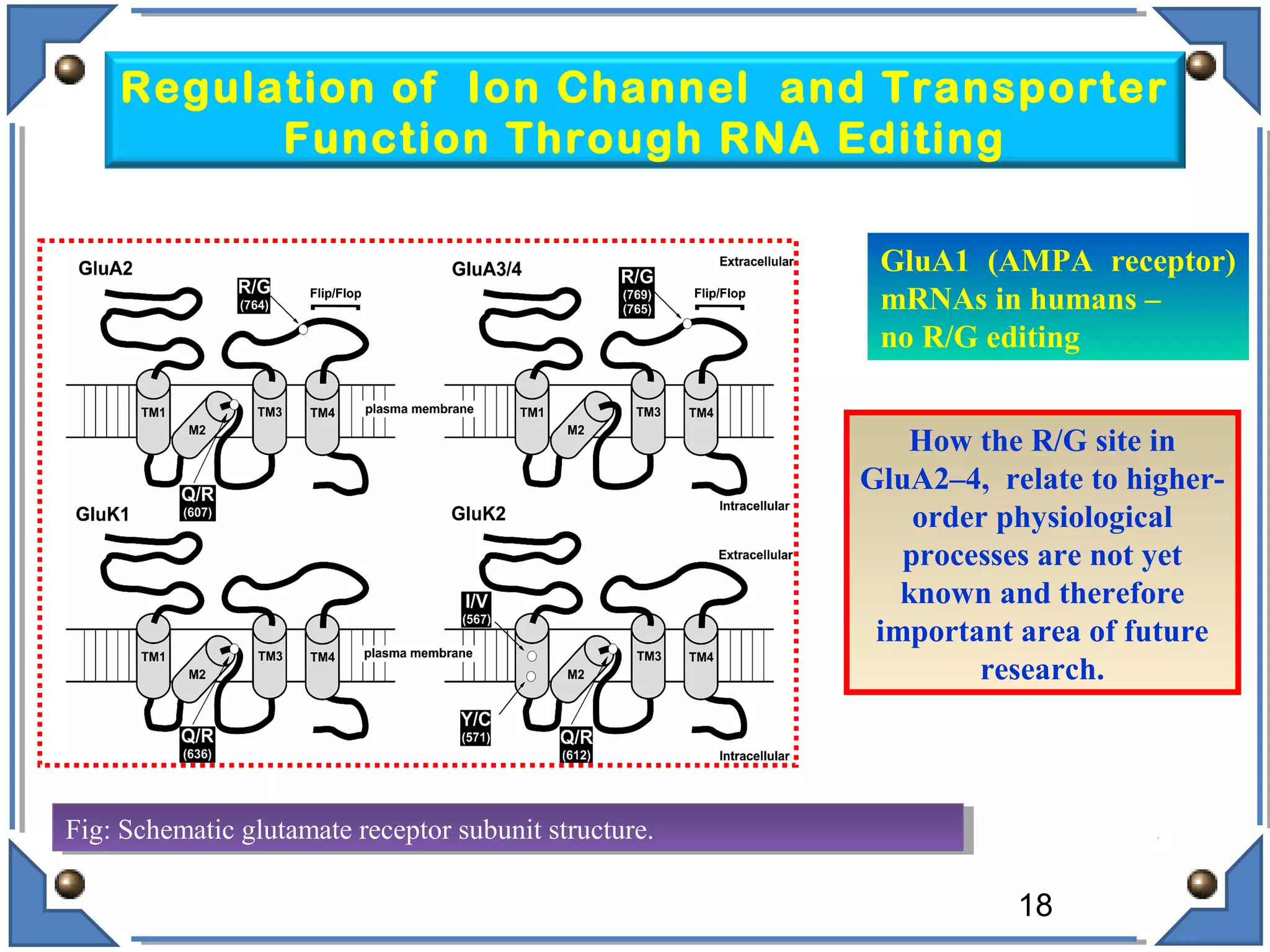
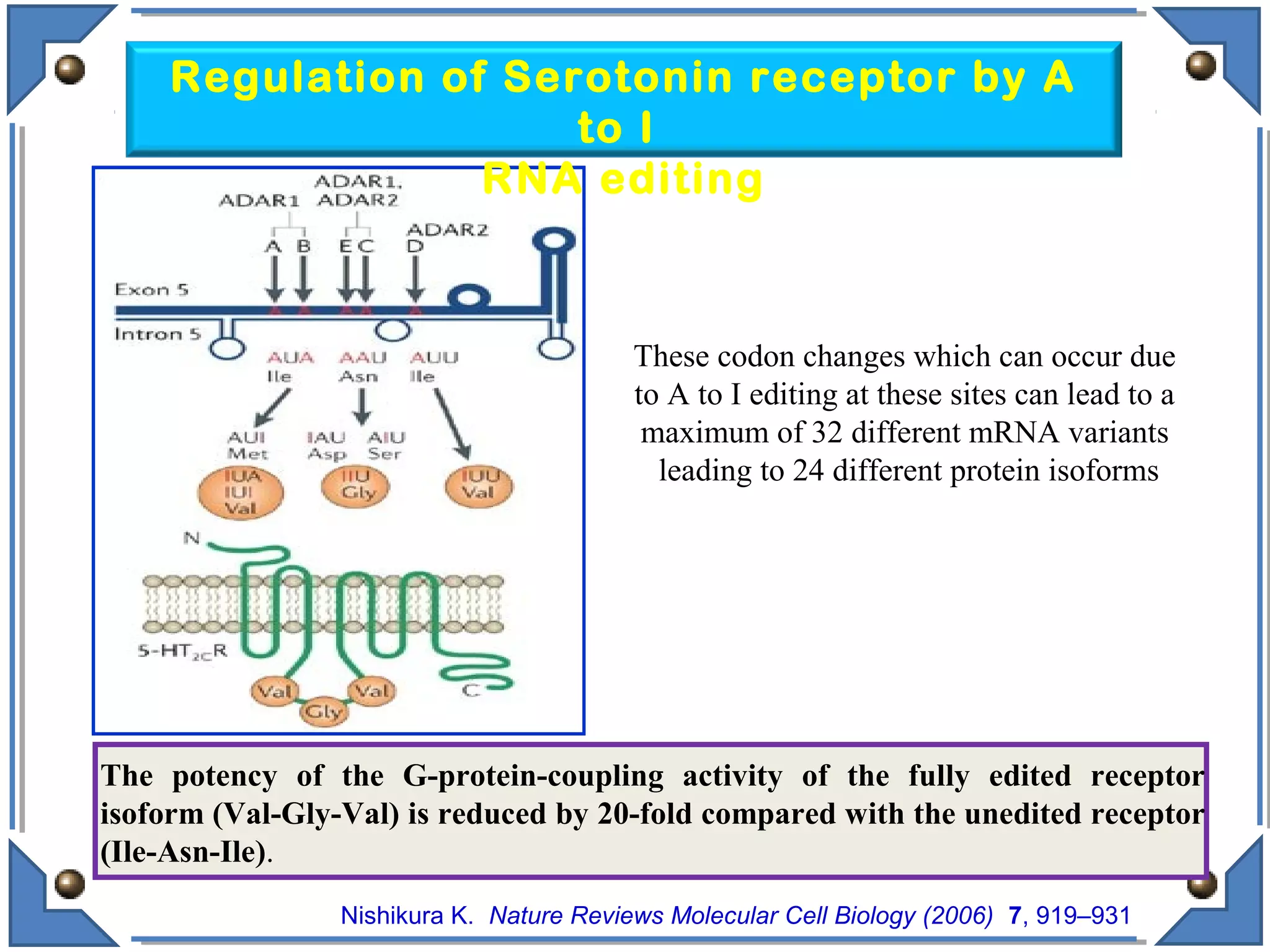
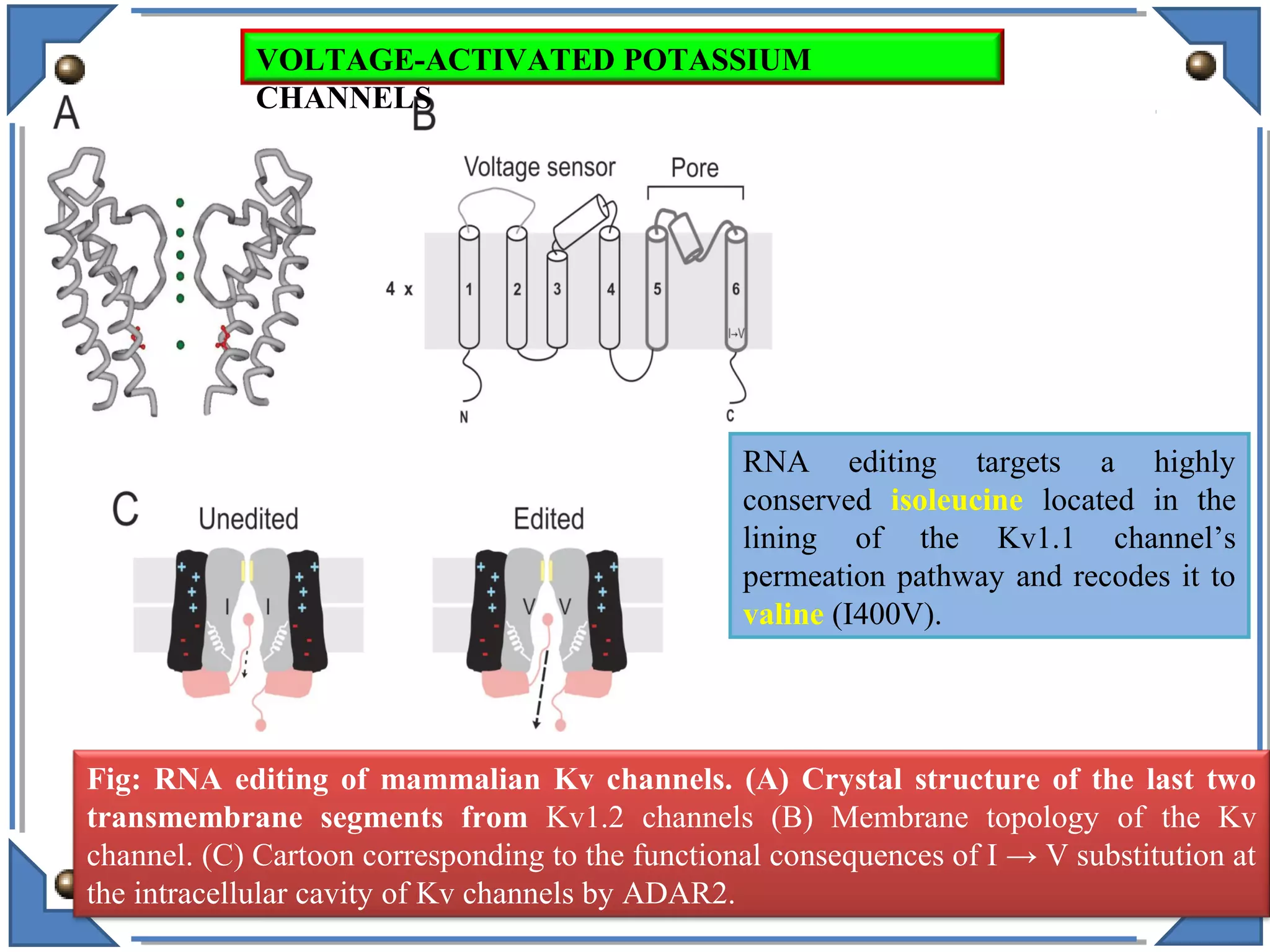
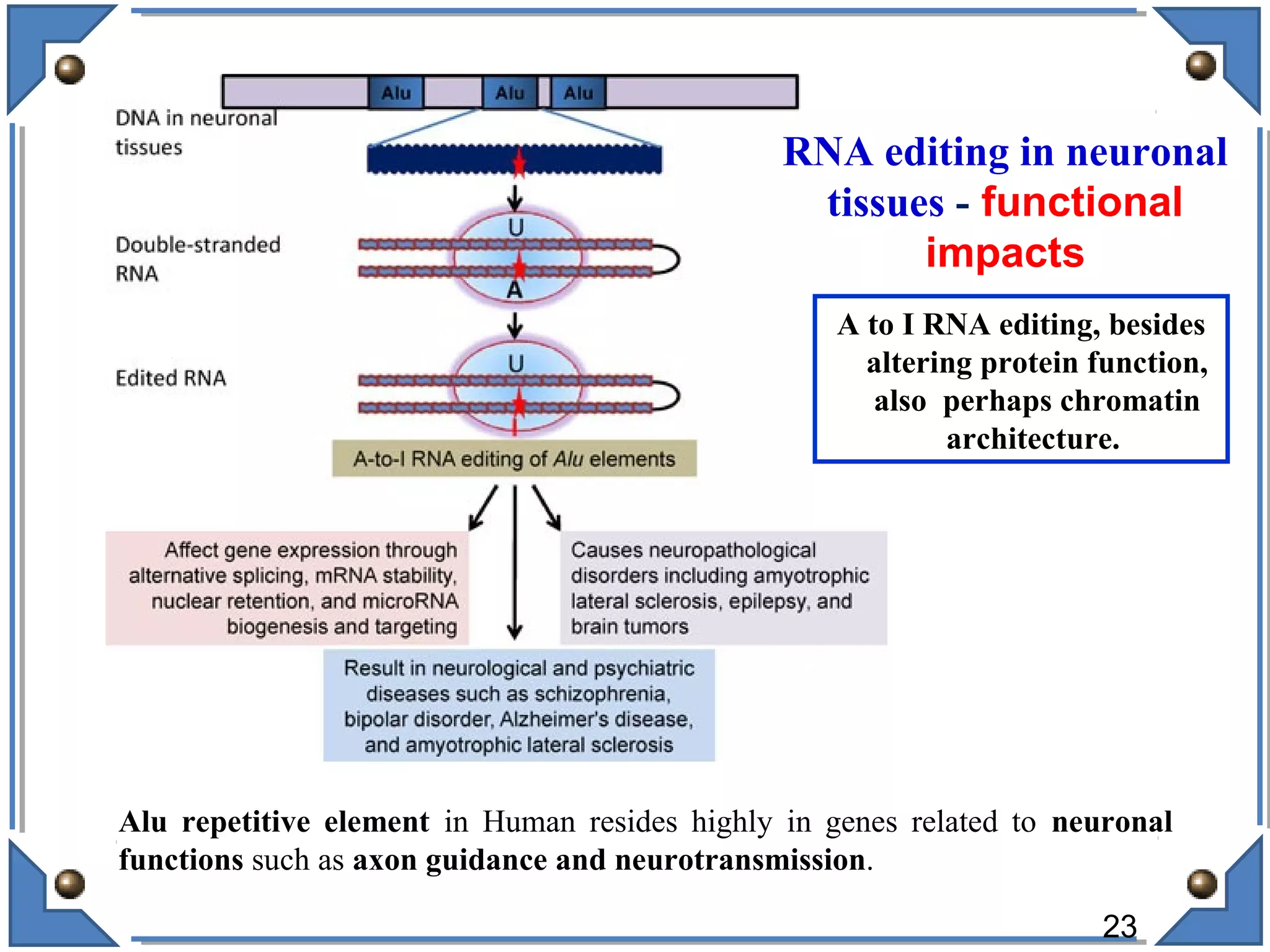
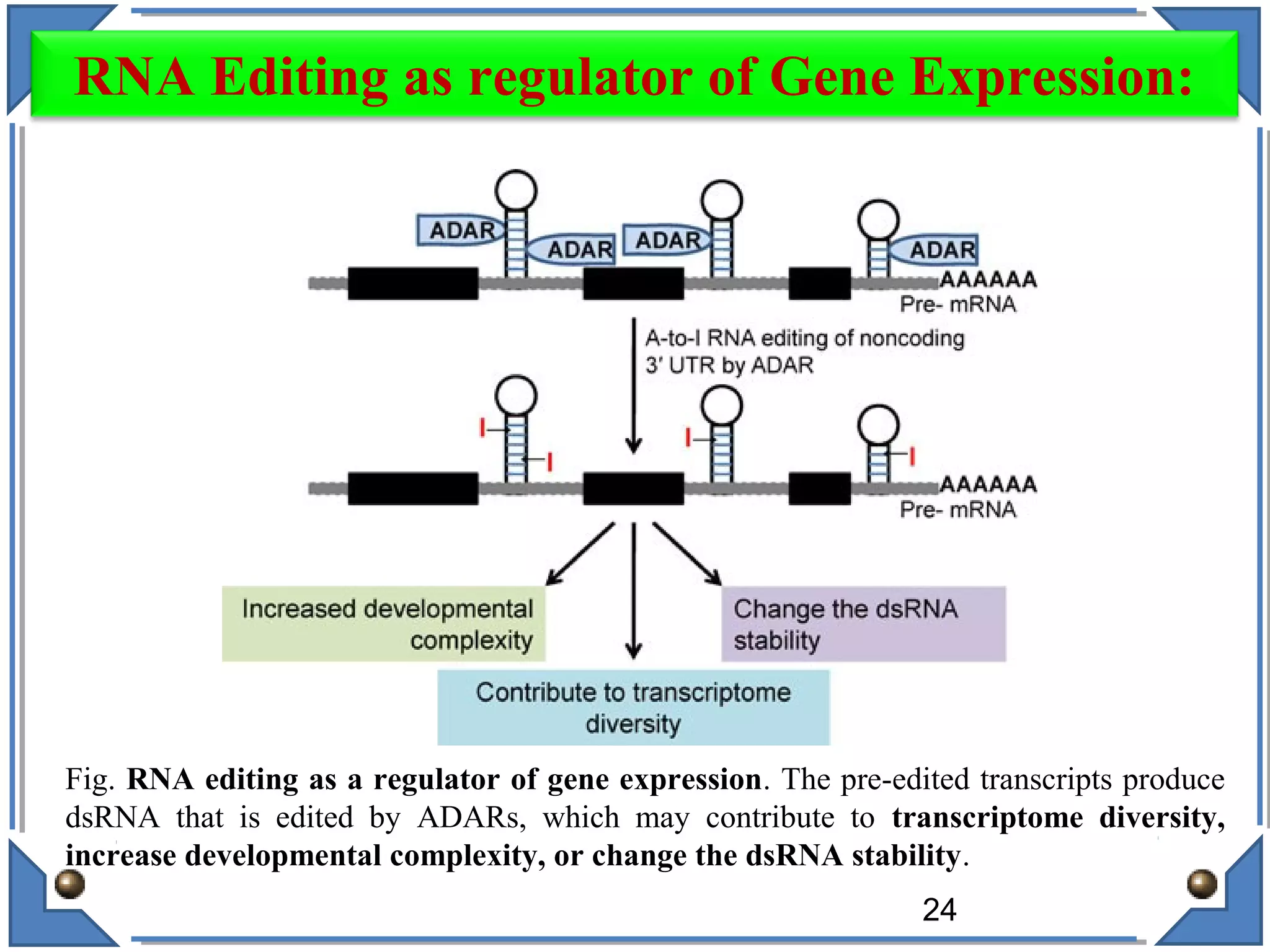
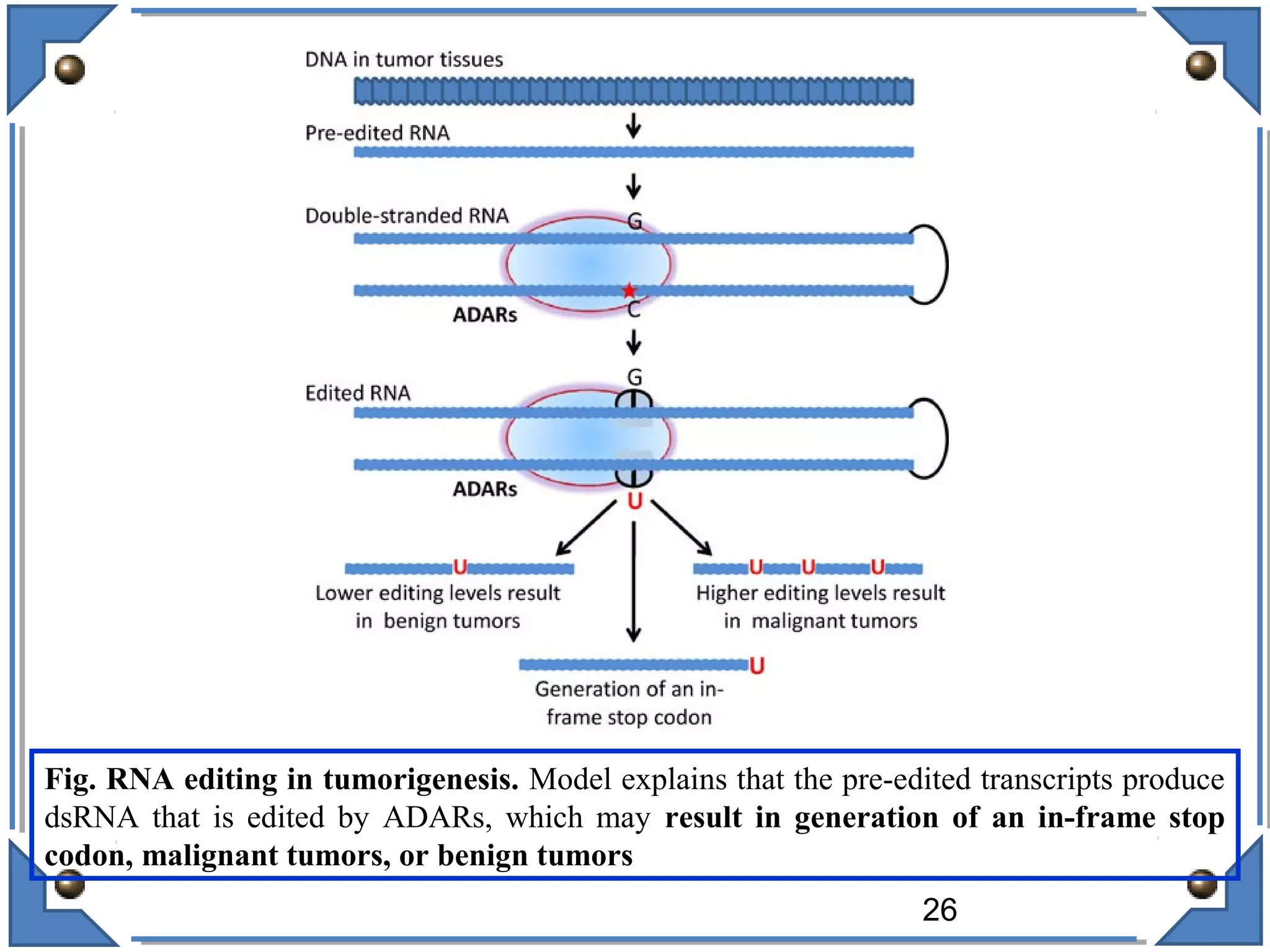
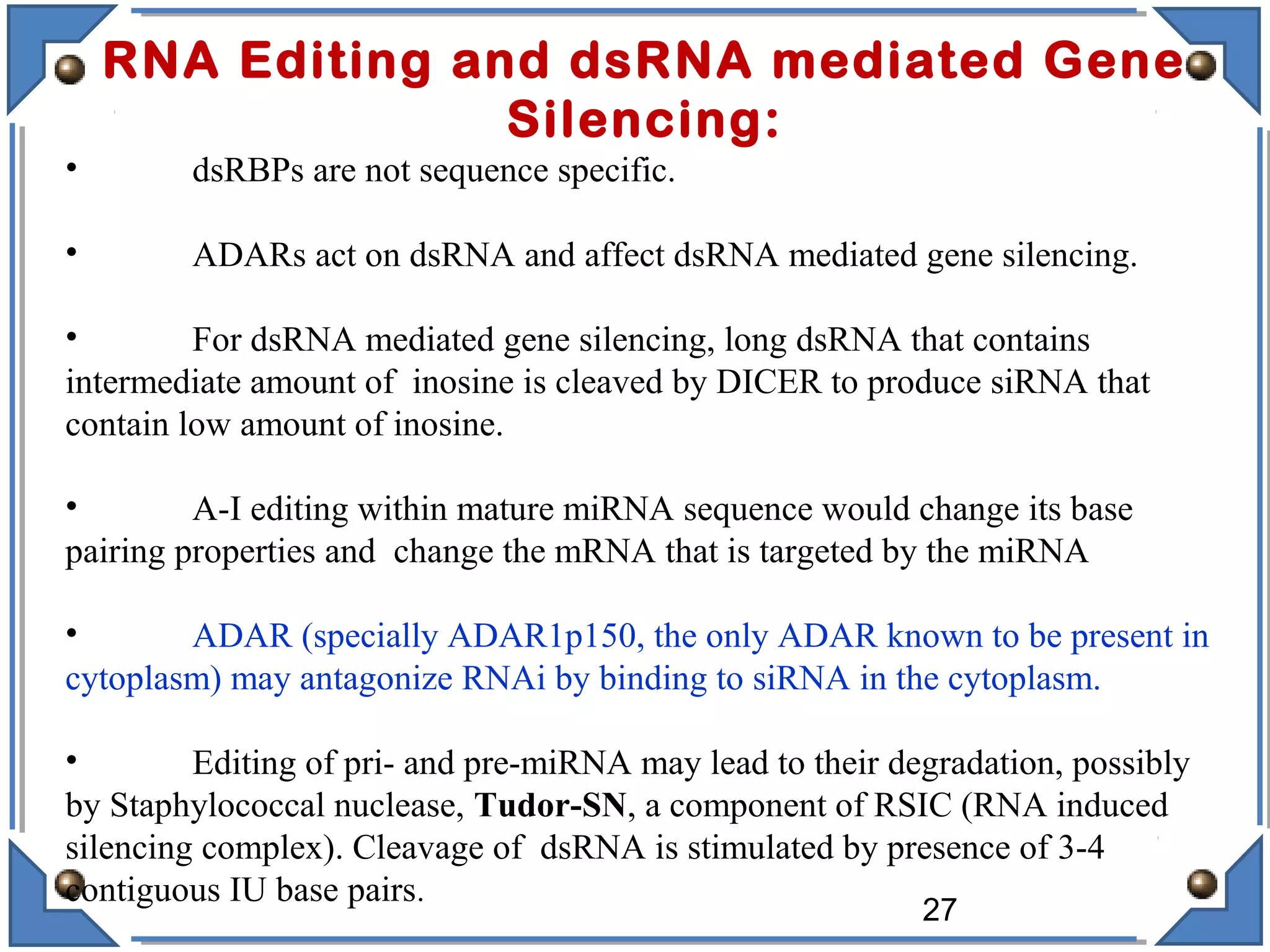
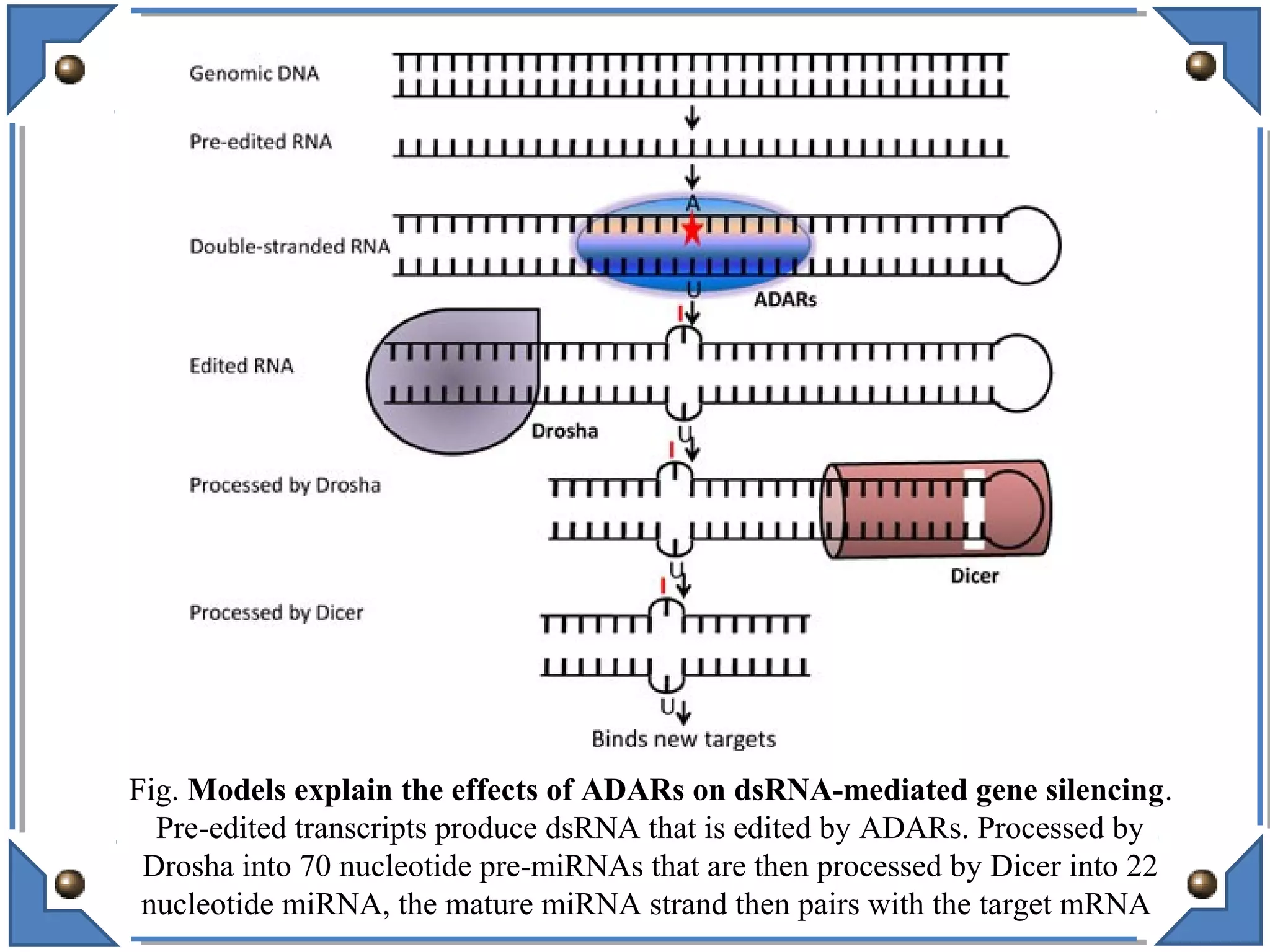
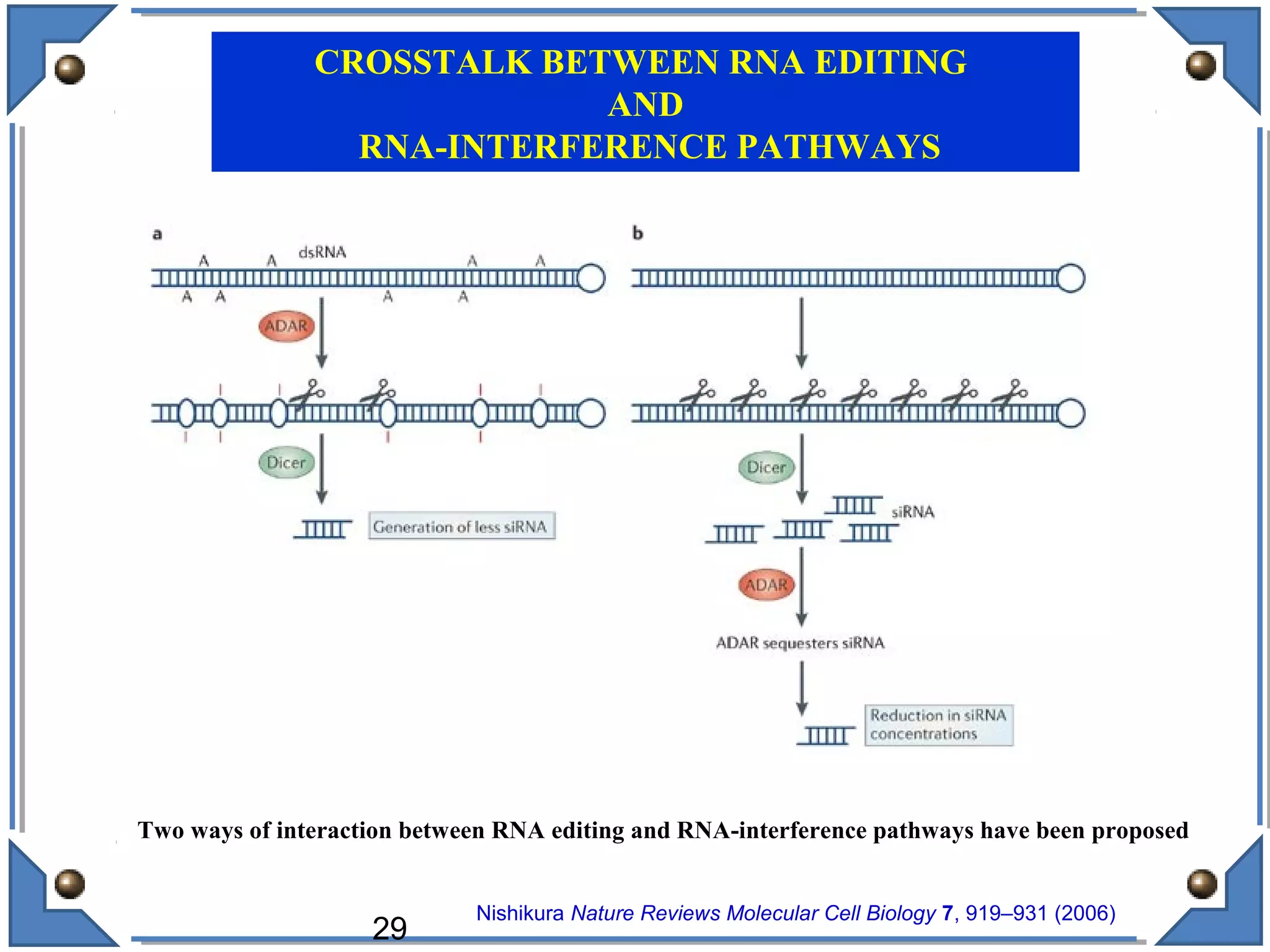
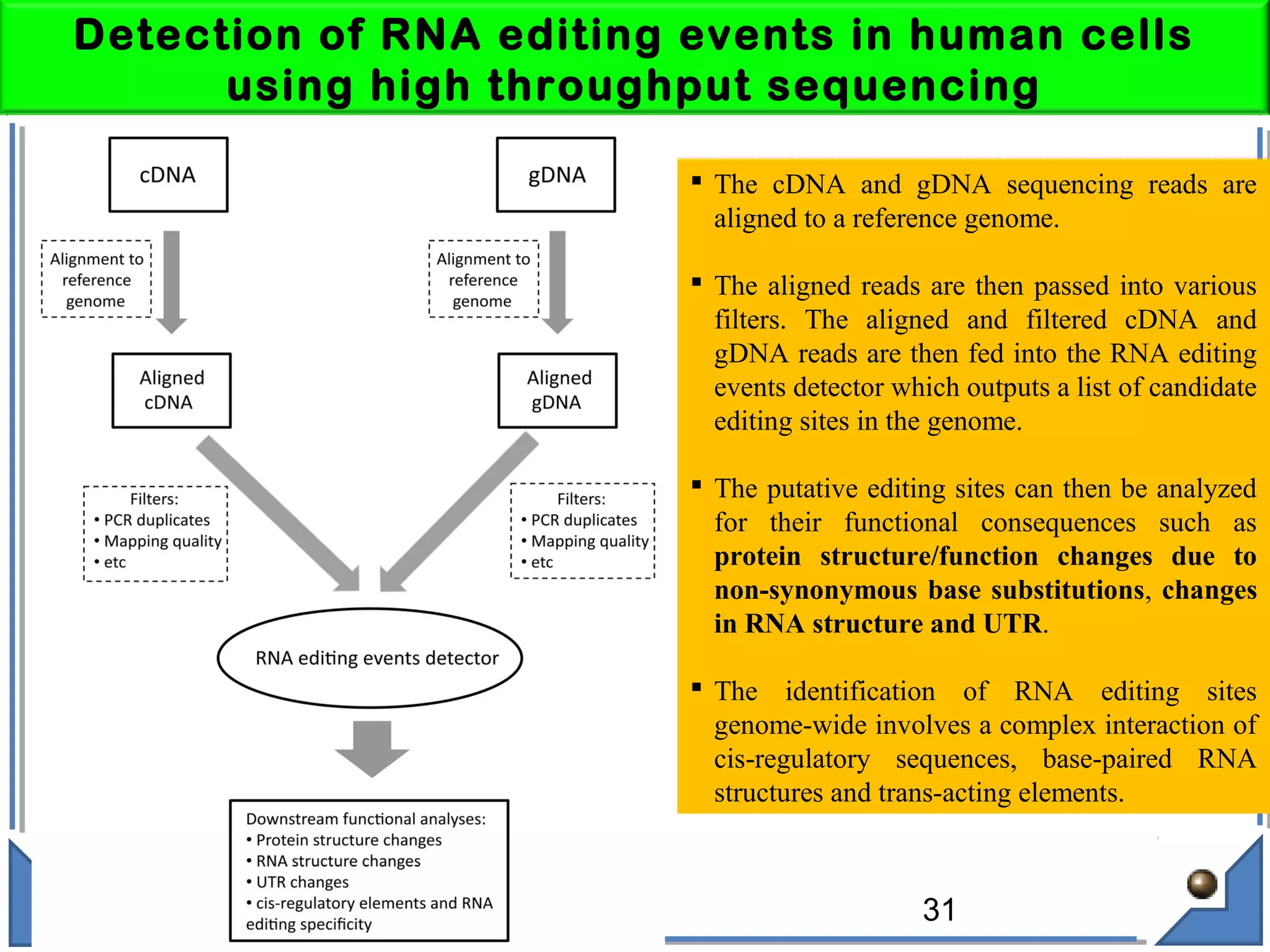
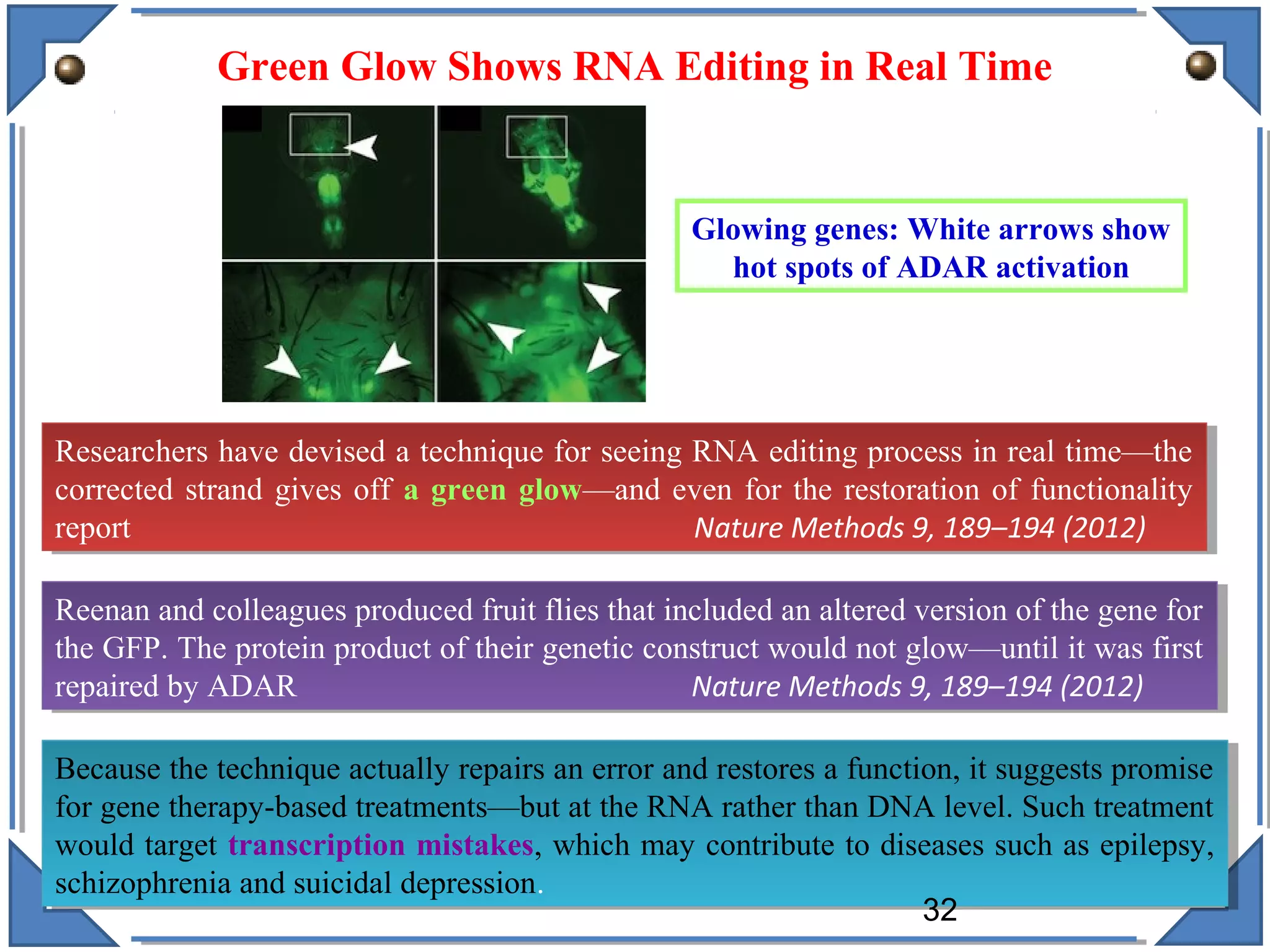
The document discusses RNA editing's role in gene expression and its potential as a therapeutic target in diseases. It outlines RNA processing mechanisms including splicing, deamination, and the involvement of various types of RNA, such as small non-coding and long non-coding RNAs in epigenetic regulation. Additionally, the document highlights the implications of RNA editing in neurological functions and tumorigenesis, emphasizing its significance in human health and disease.