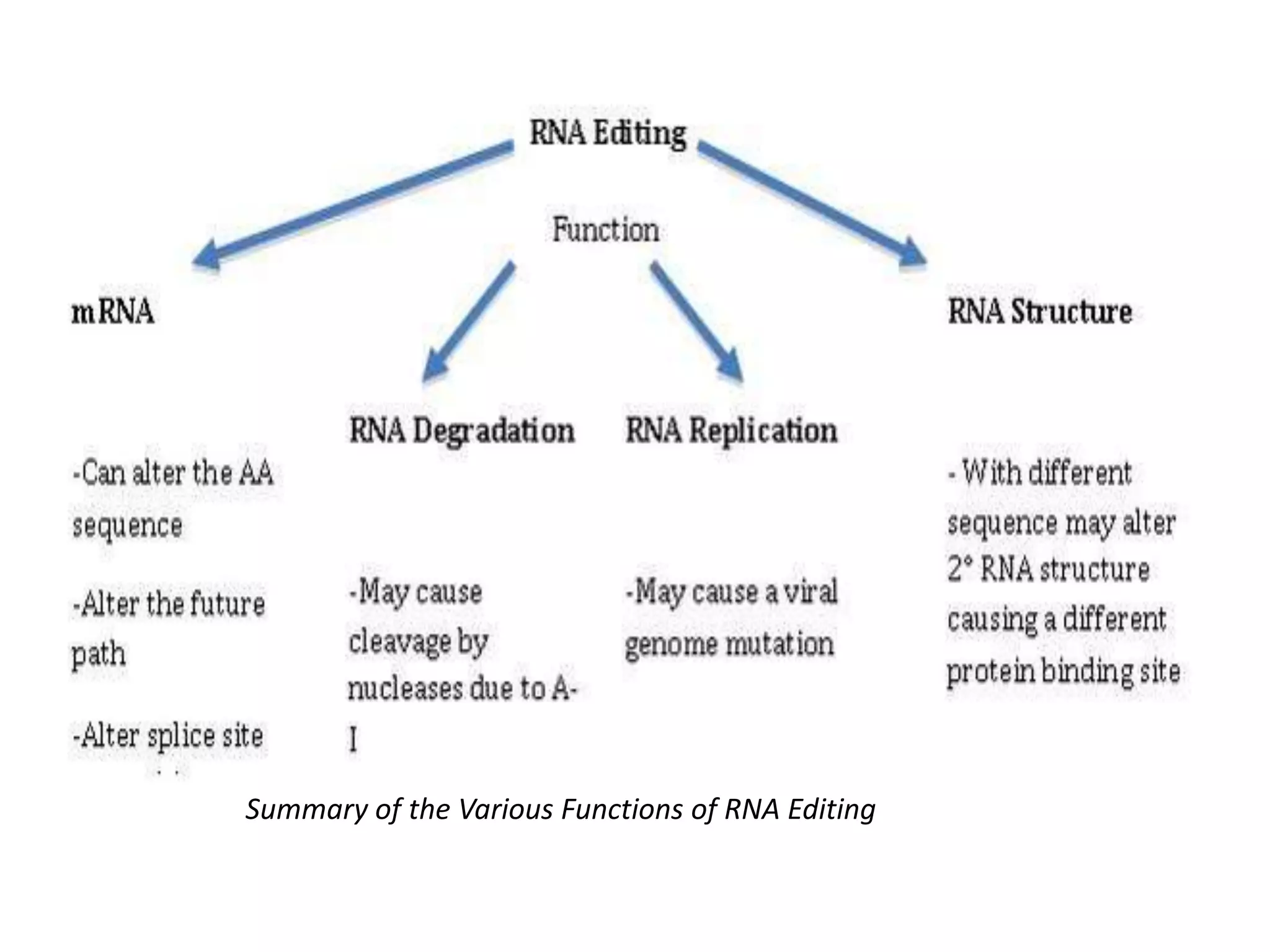
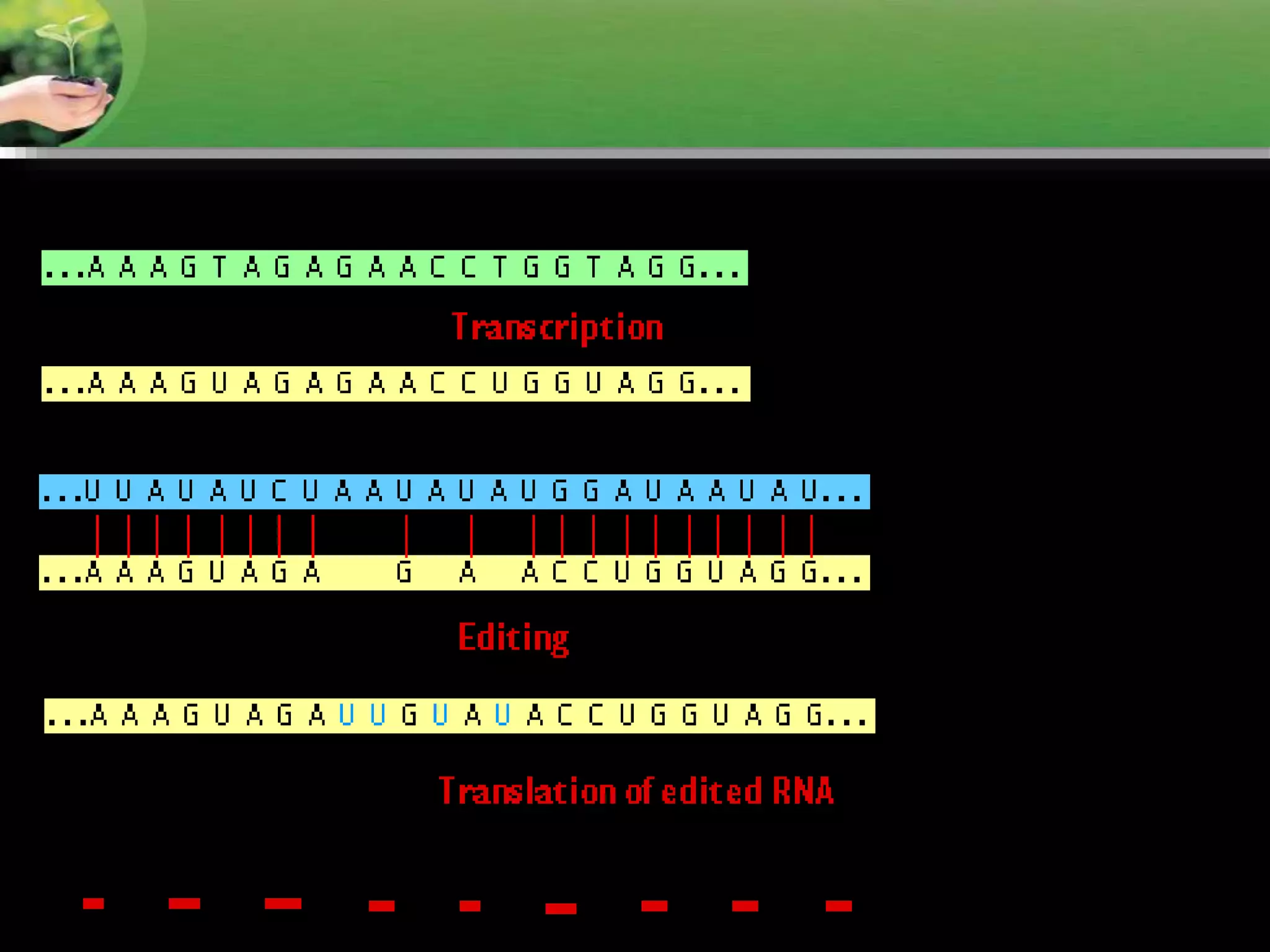
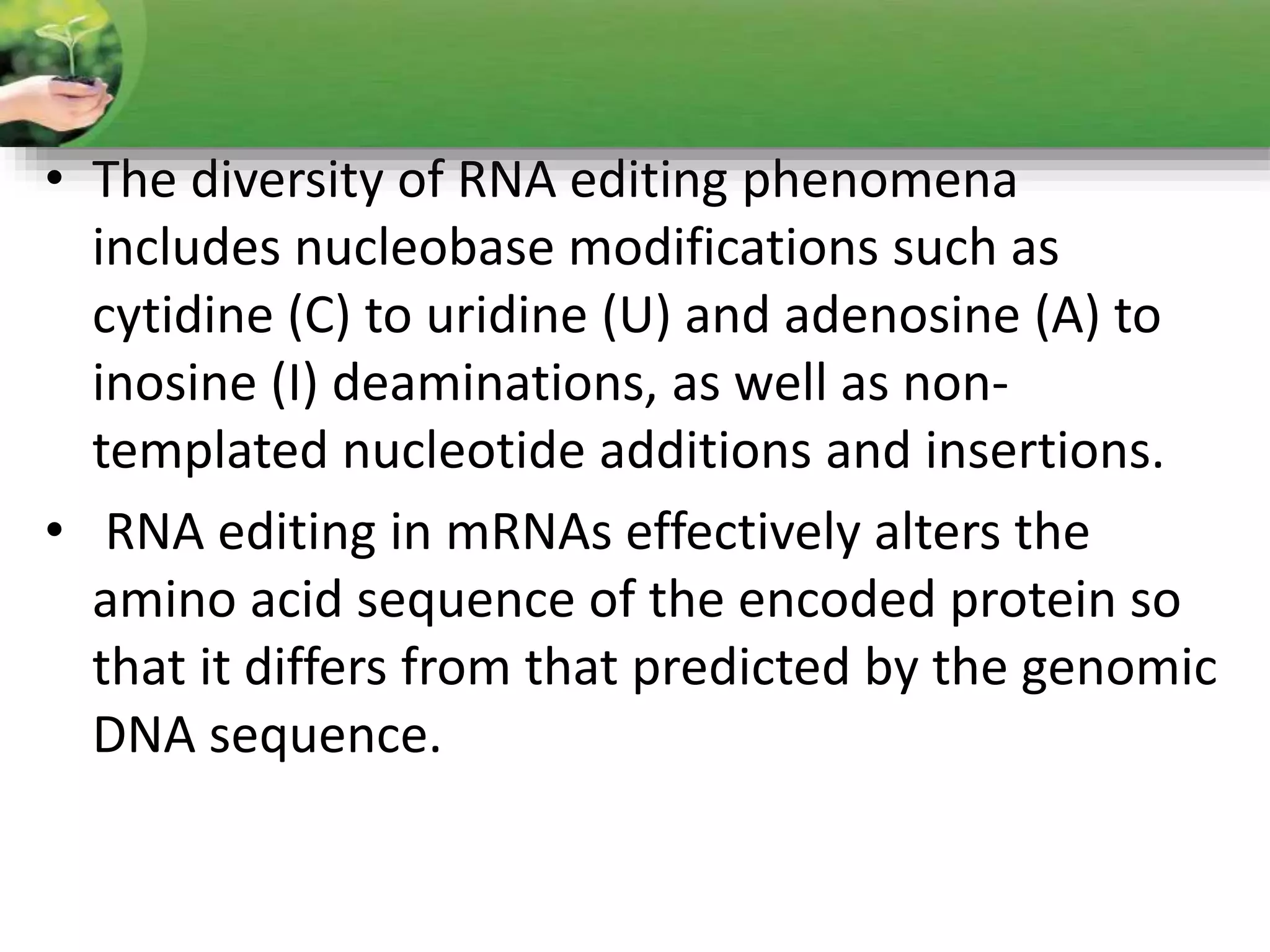
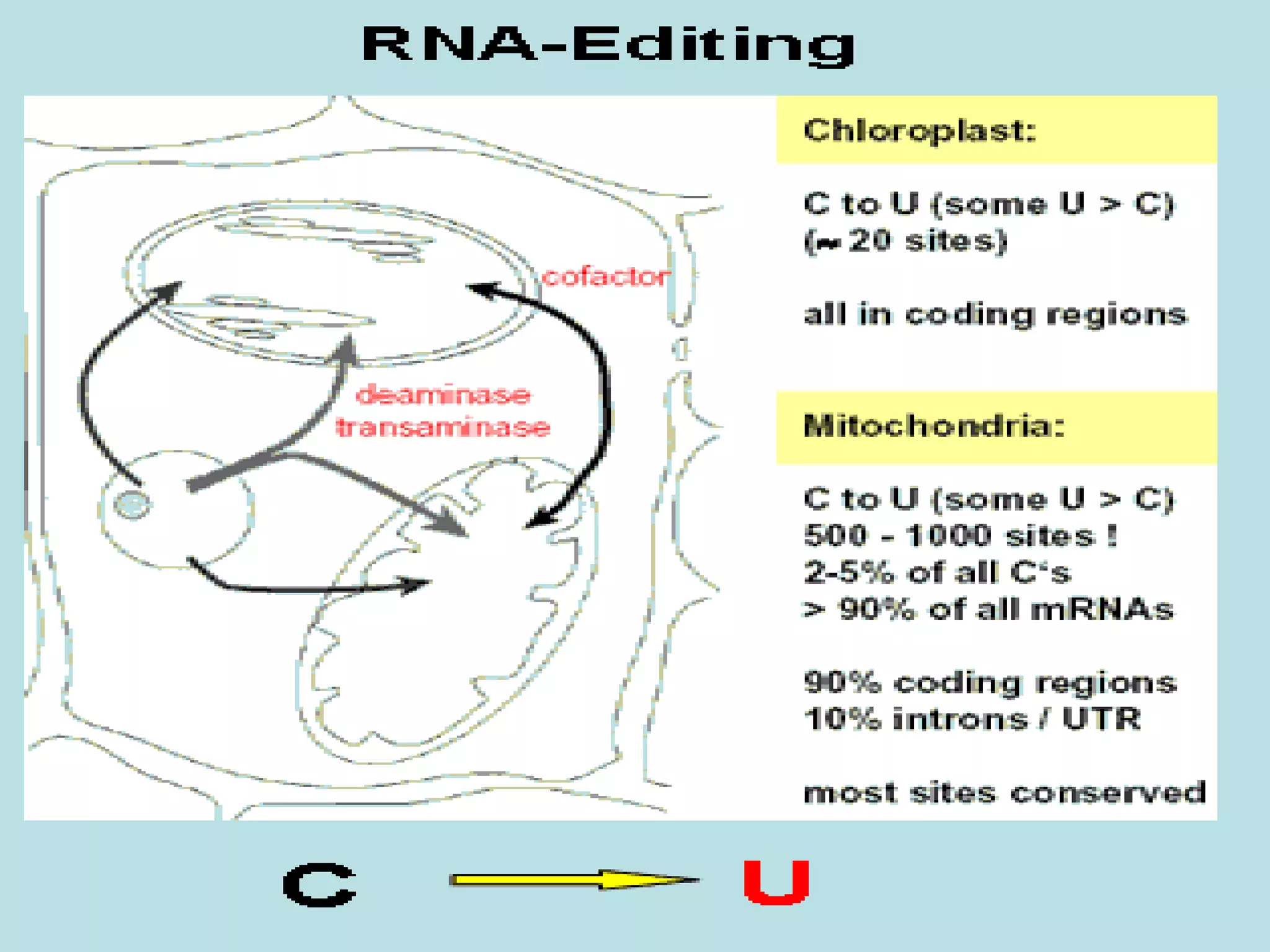
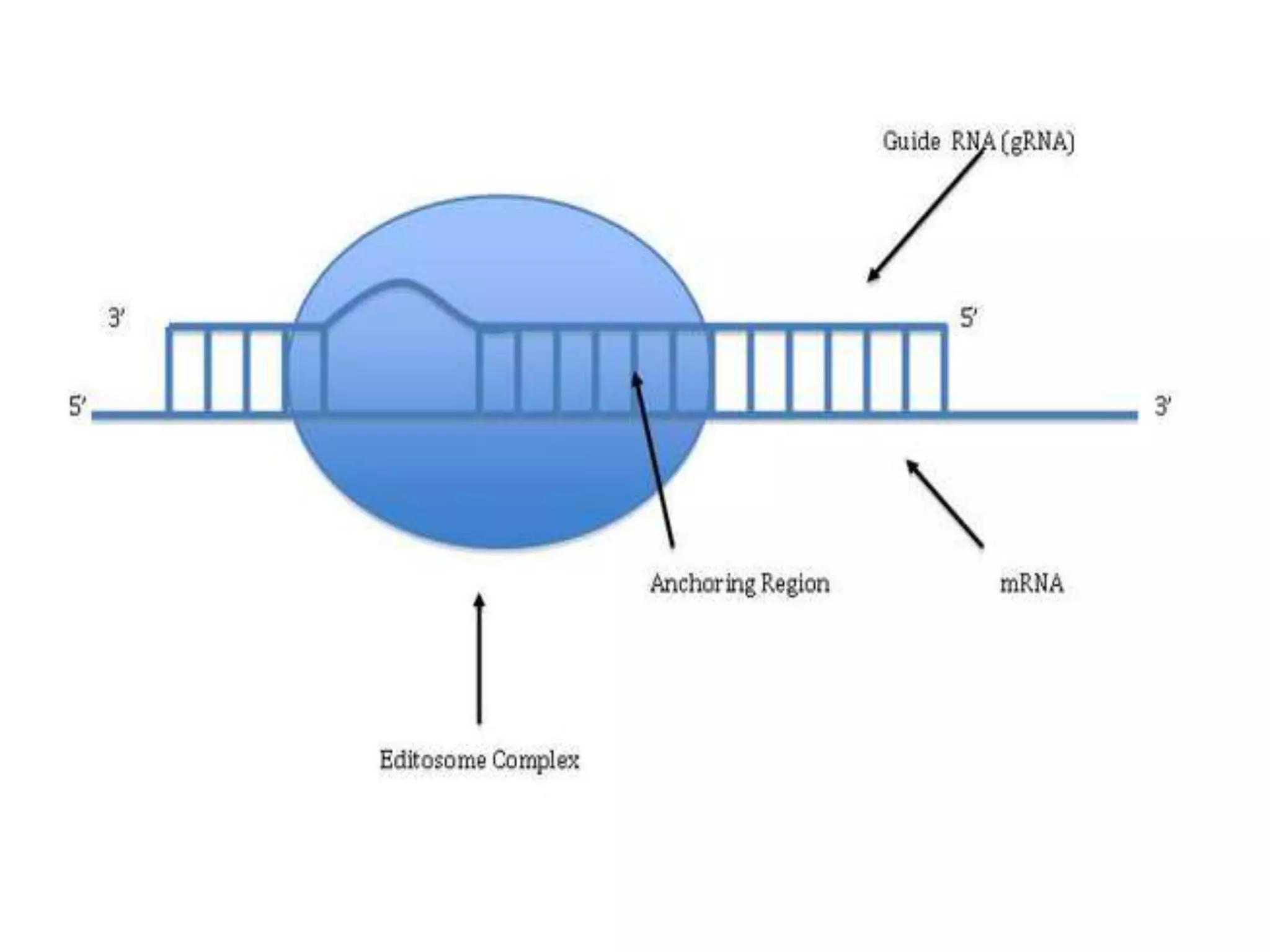
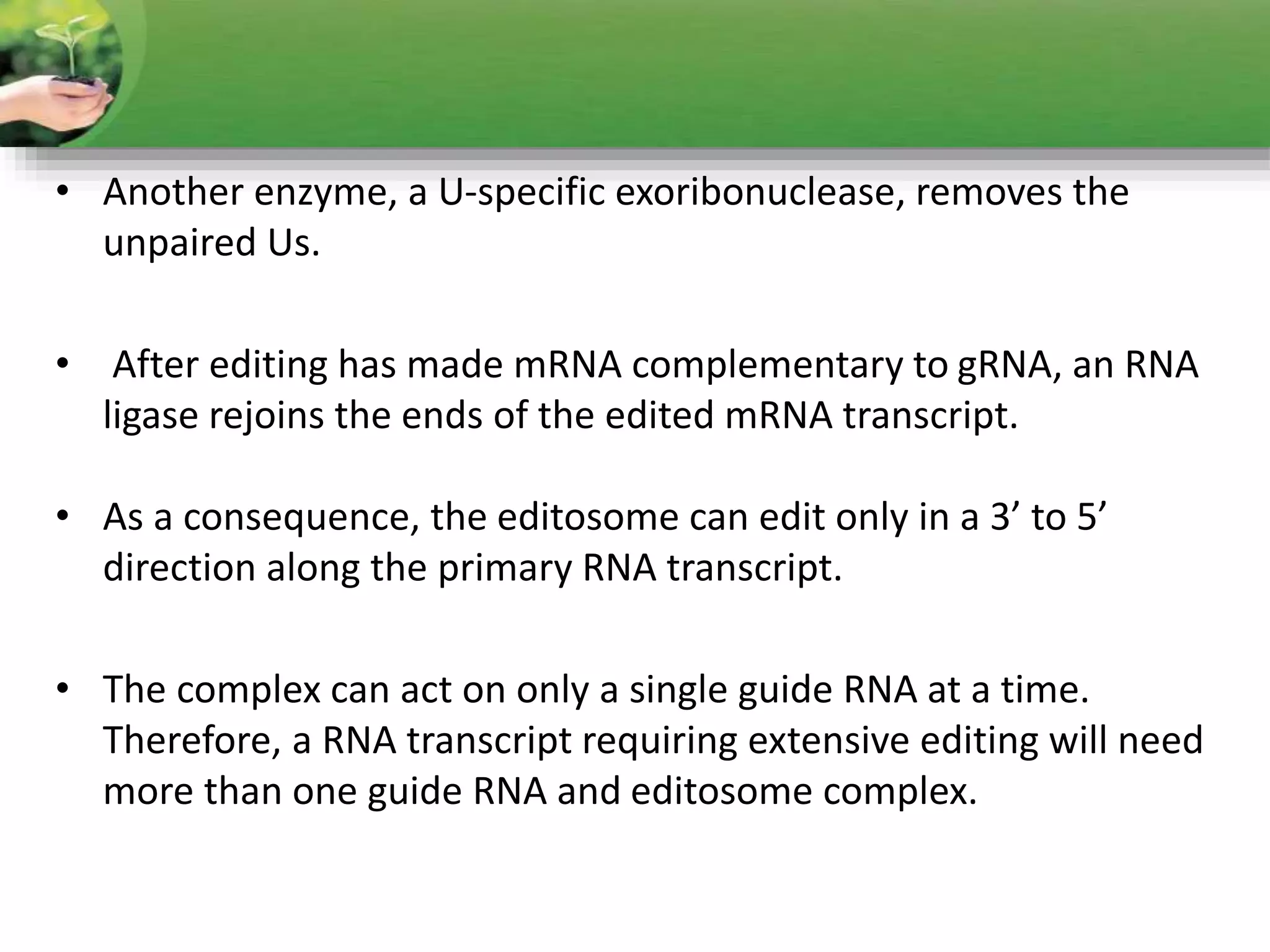
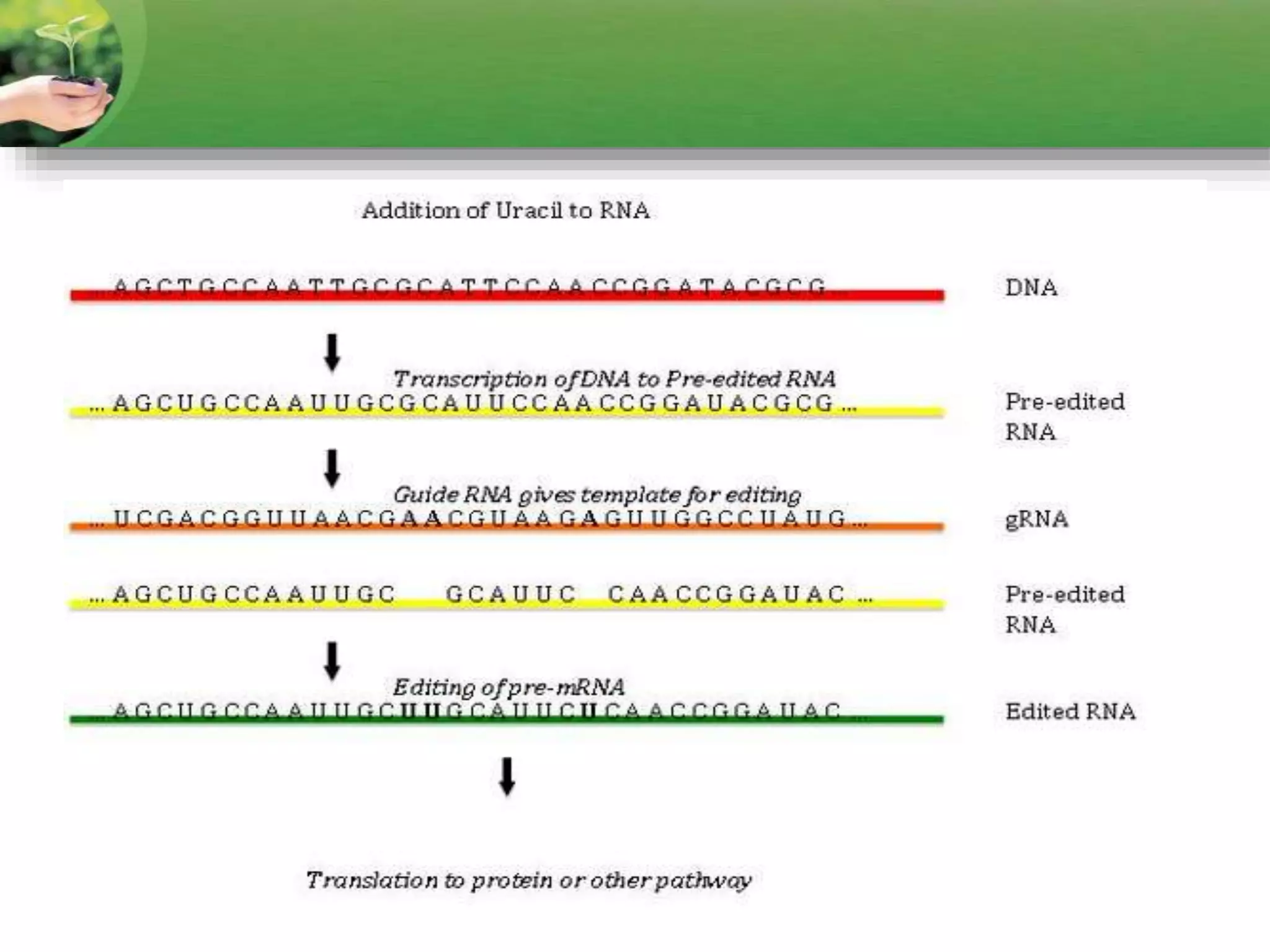
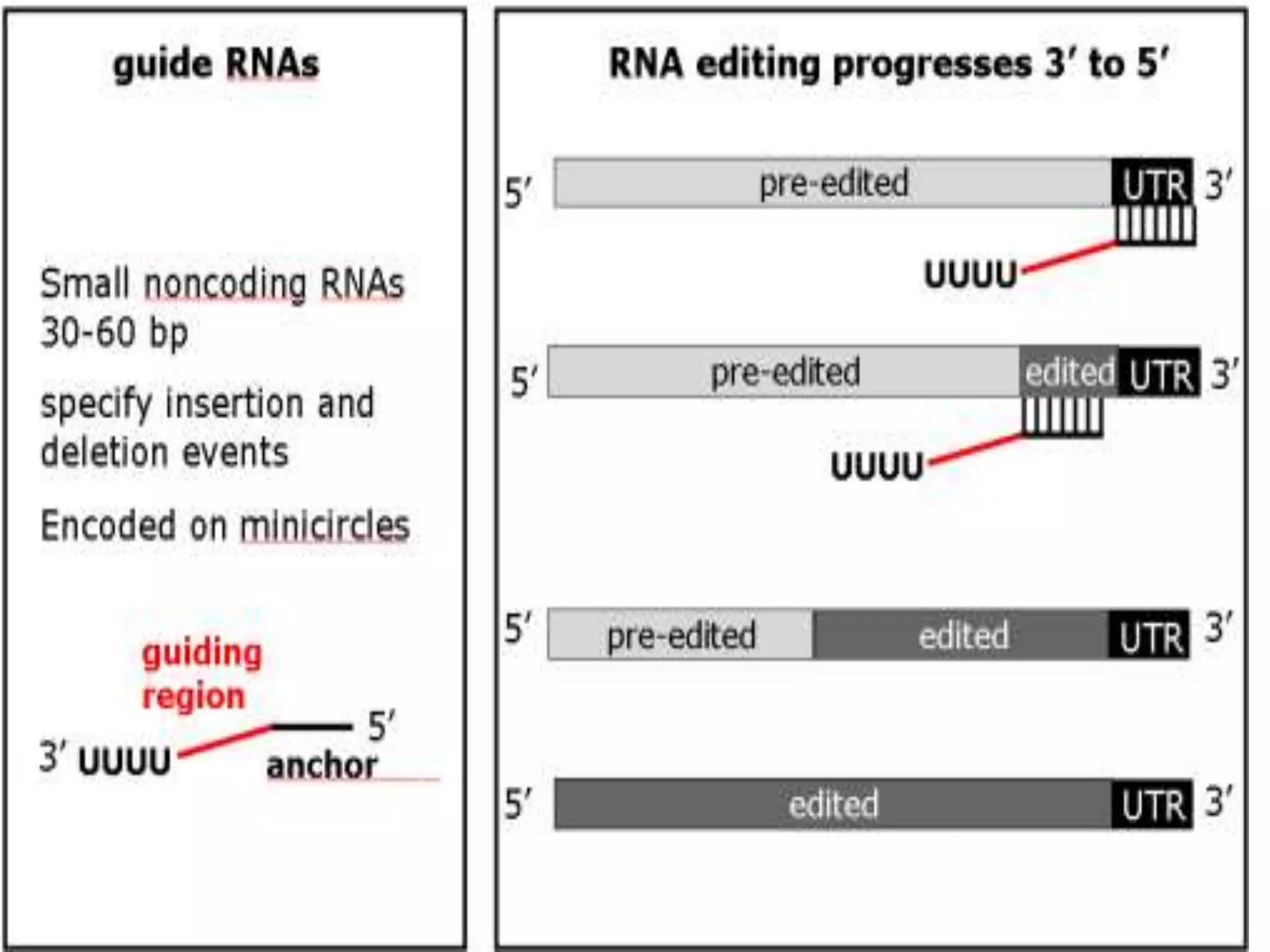
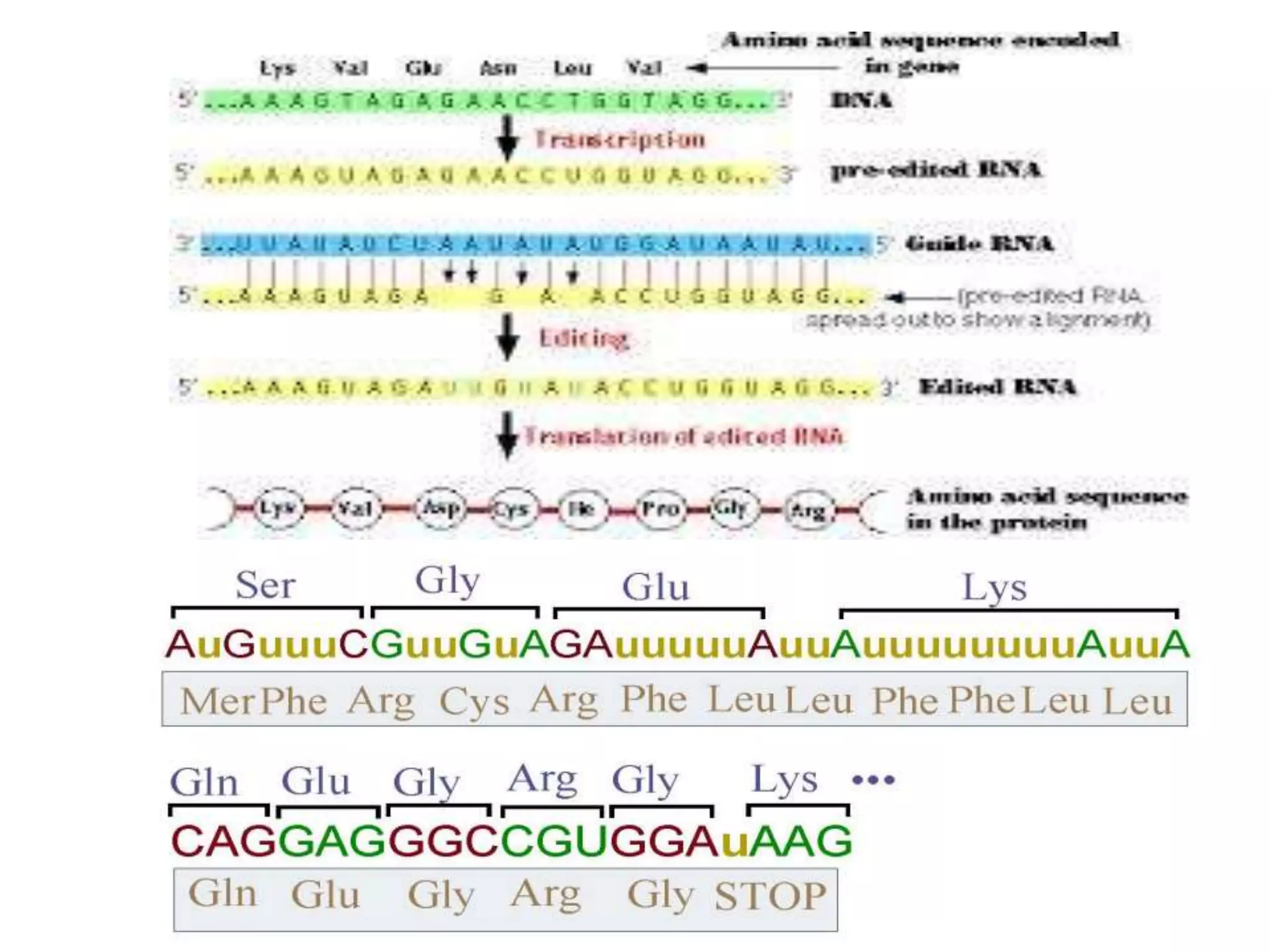
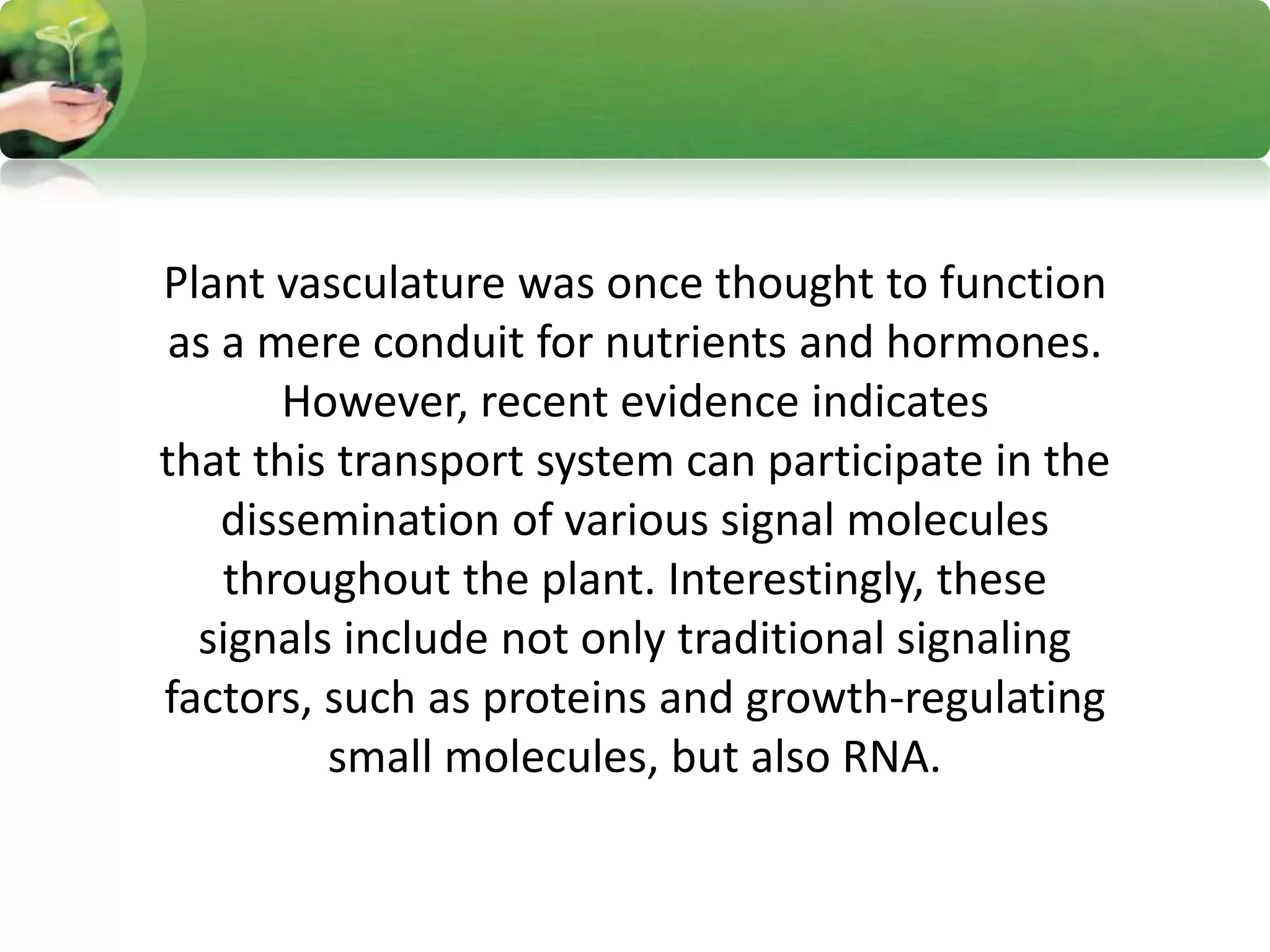
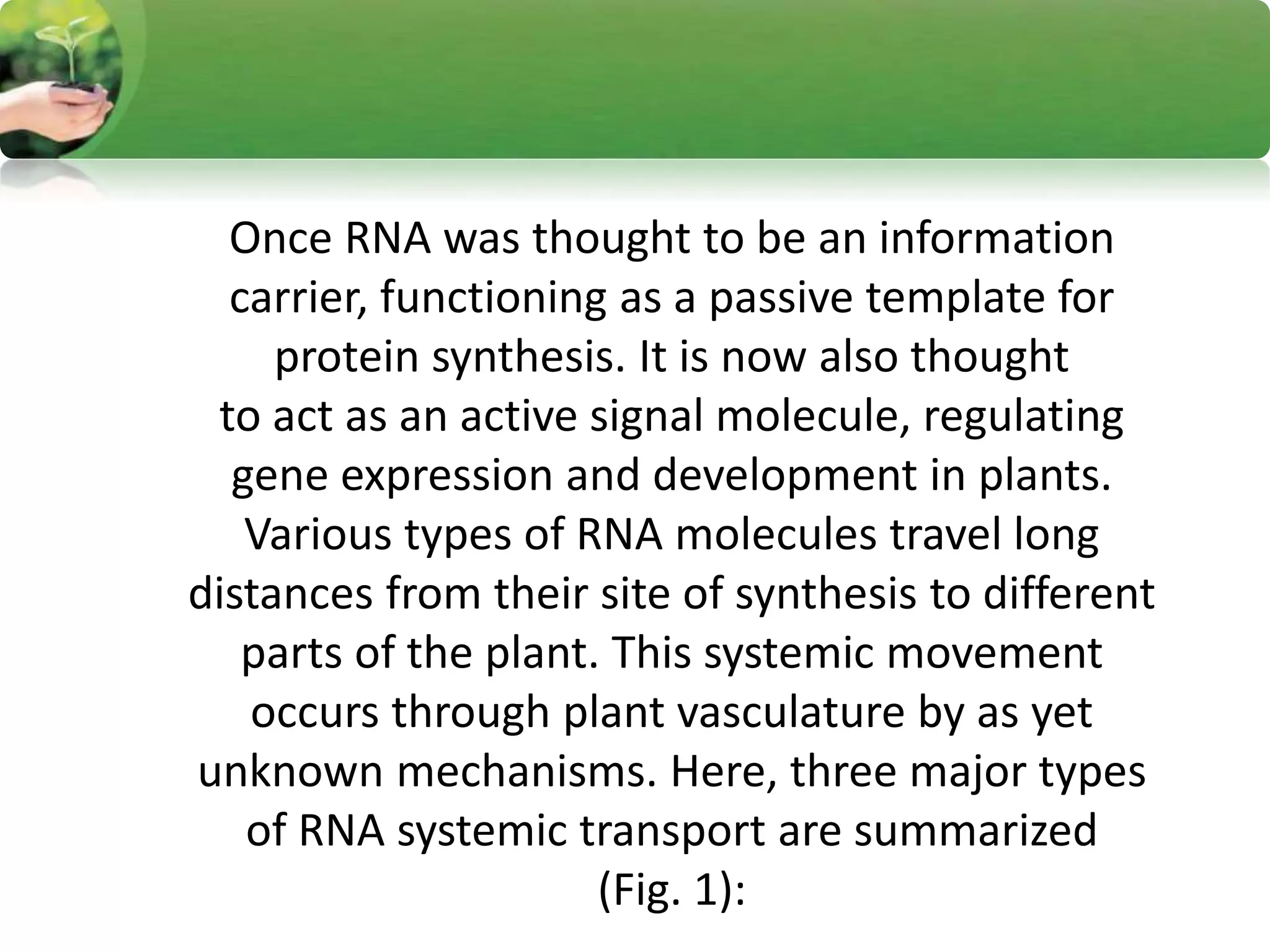
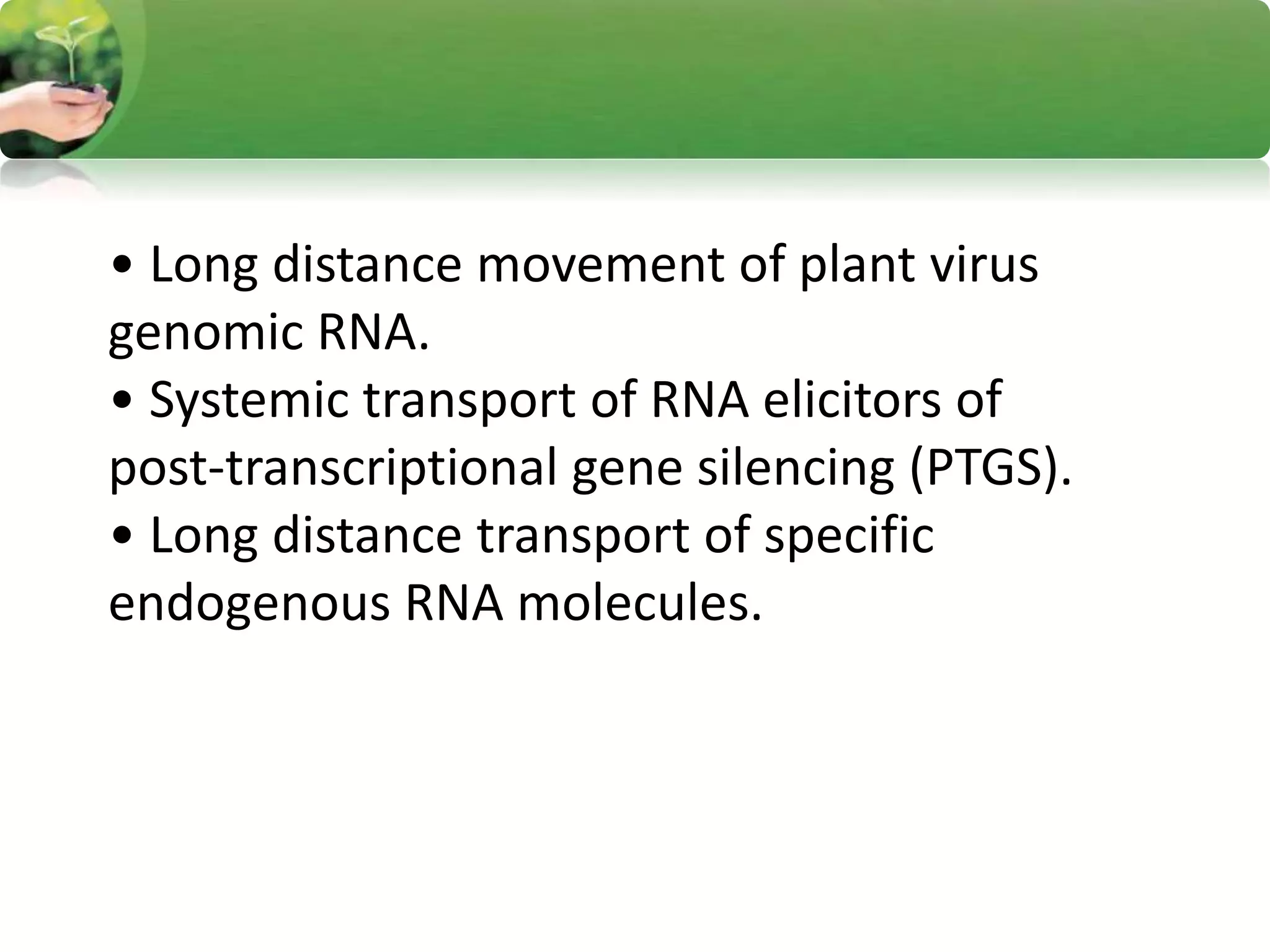
- RNA editing is a process where the RNA sequence differs from the DNA sequence due to modifications like base substitutions, insertions or deletions.
- RNA editing occurs in various RNA molecules like mRNA, tRNA, rRNA in eukaryotes, archaea, prokaryotes in the cell nucleus, cytosol, mitochondria and plastids.
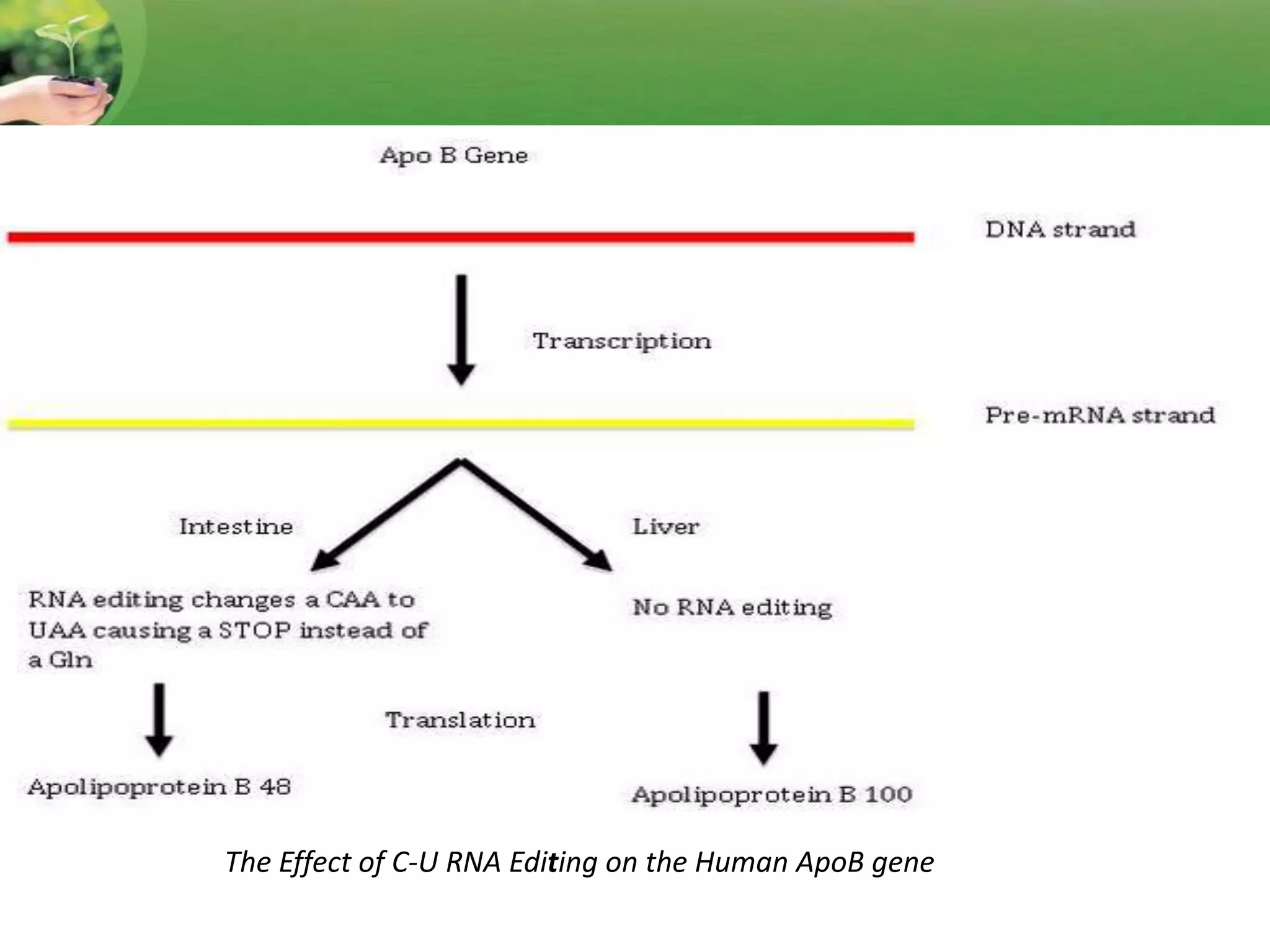
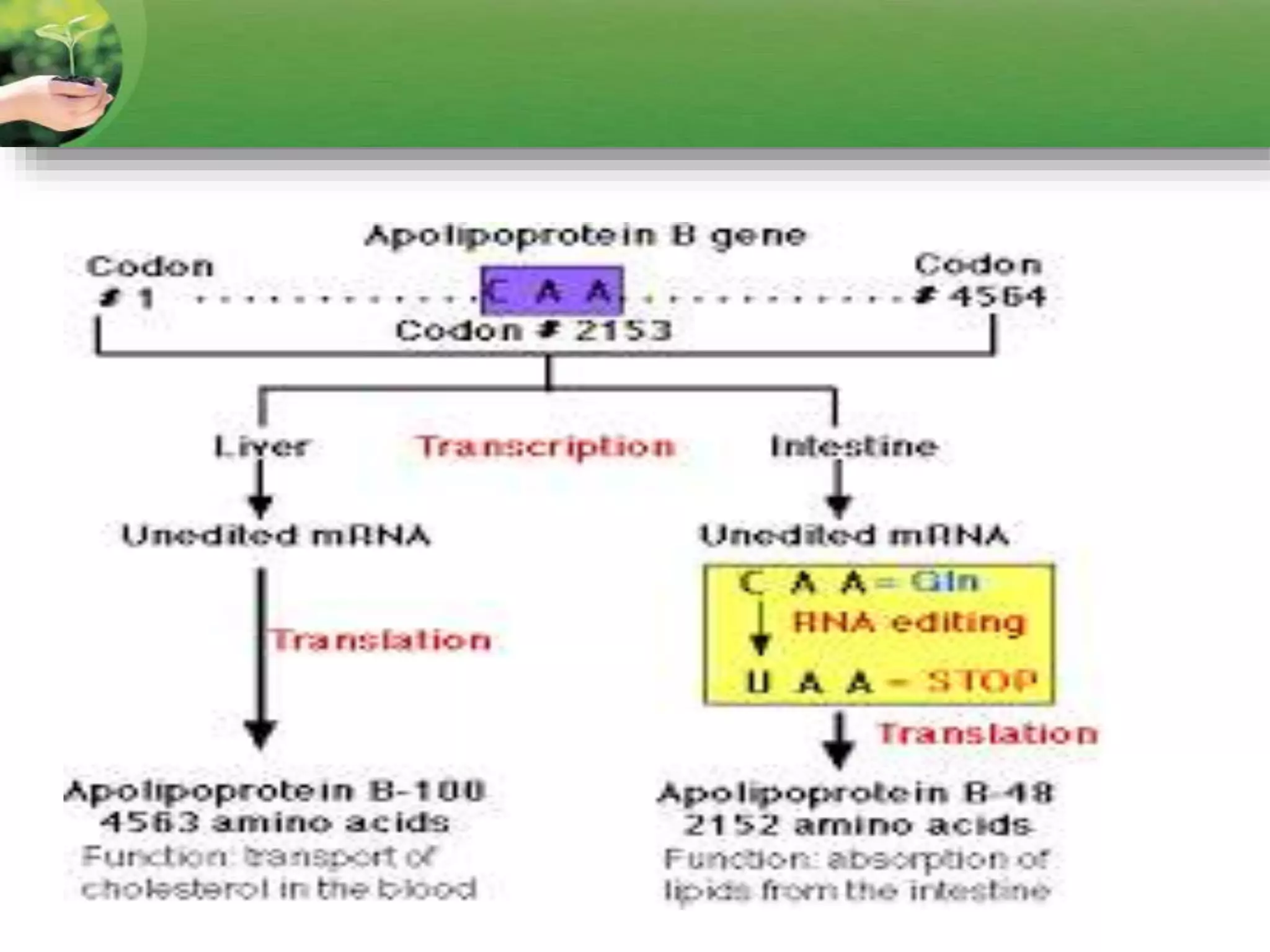
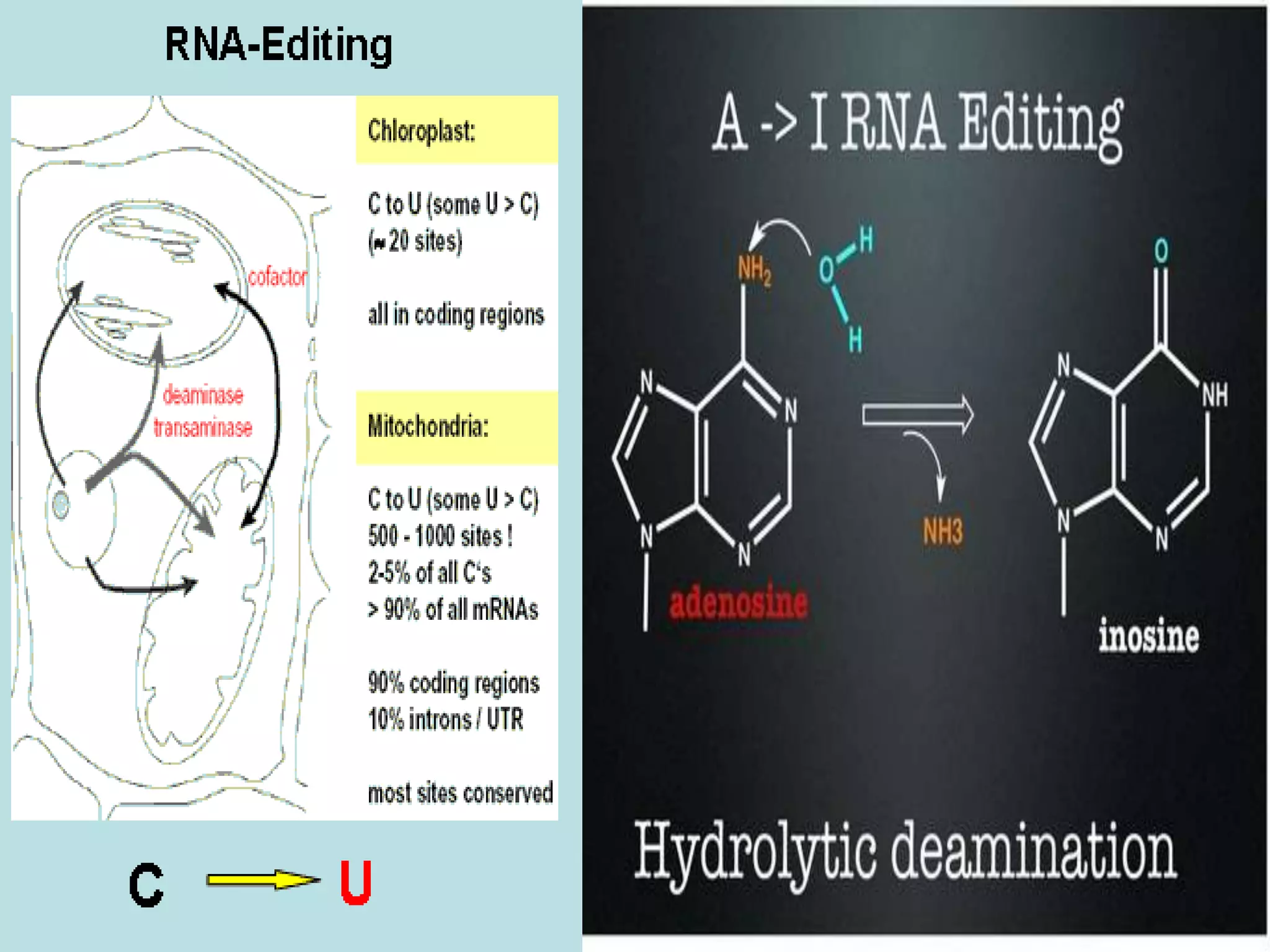
- Two major types of RNA editing are C to U editing, like in the human ApoB gene, and A to I editing which occurs in double-stranded RNA regions and is catalyzed by ADAR enzymes.