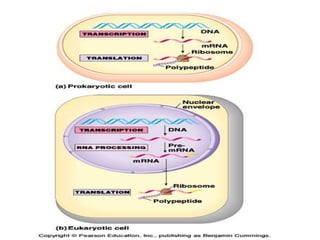
The document discusses various post-transcriptional processes that eukaryotic RNA undergo. It describes:
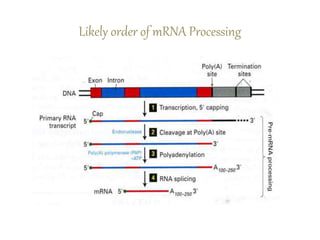
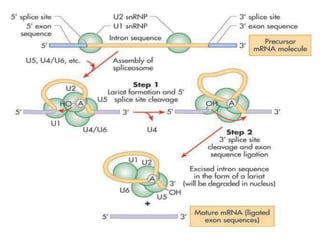
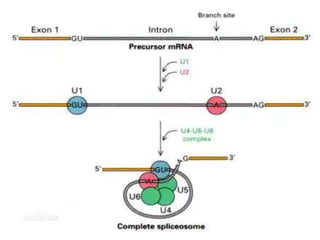
1) Introns are removed and exons are joined together in mRNA through splicing. Small nuclear RNAs and spliceosomes are involved in splicing.
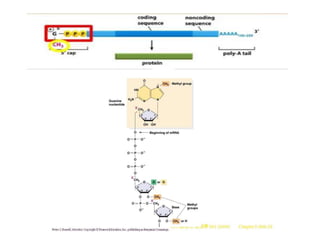
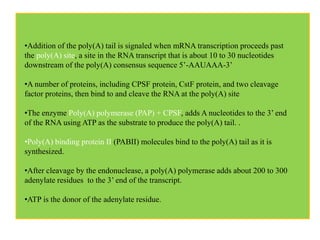
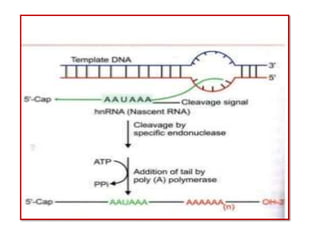
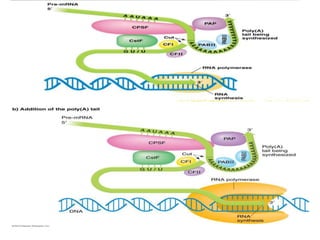
2) Other processing includes adding a 5' cap and 3' poly-A tail. The tail protects the mRNA and is important for translation.
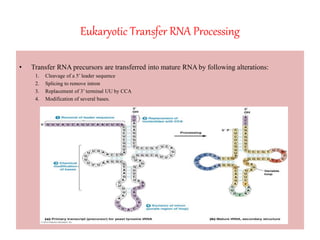
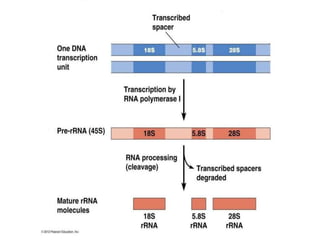
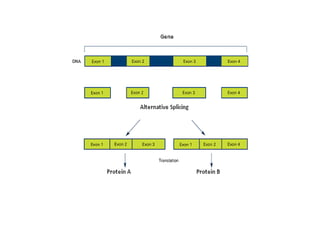
3) Transfer RNA and ribosomal RNA also undergo processing after transcription to become functional in the cell. Alternative splicing allows different mRNAs and proteins to be produced from the same initial RNA.