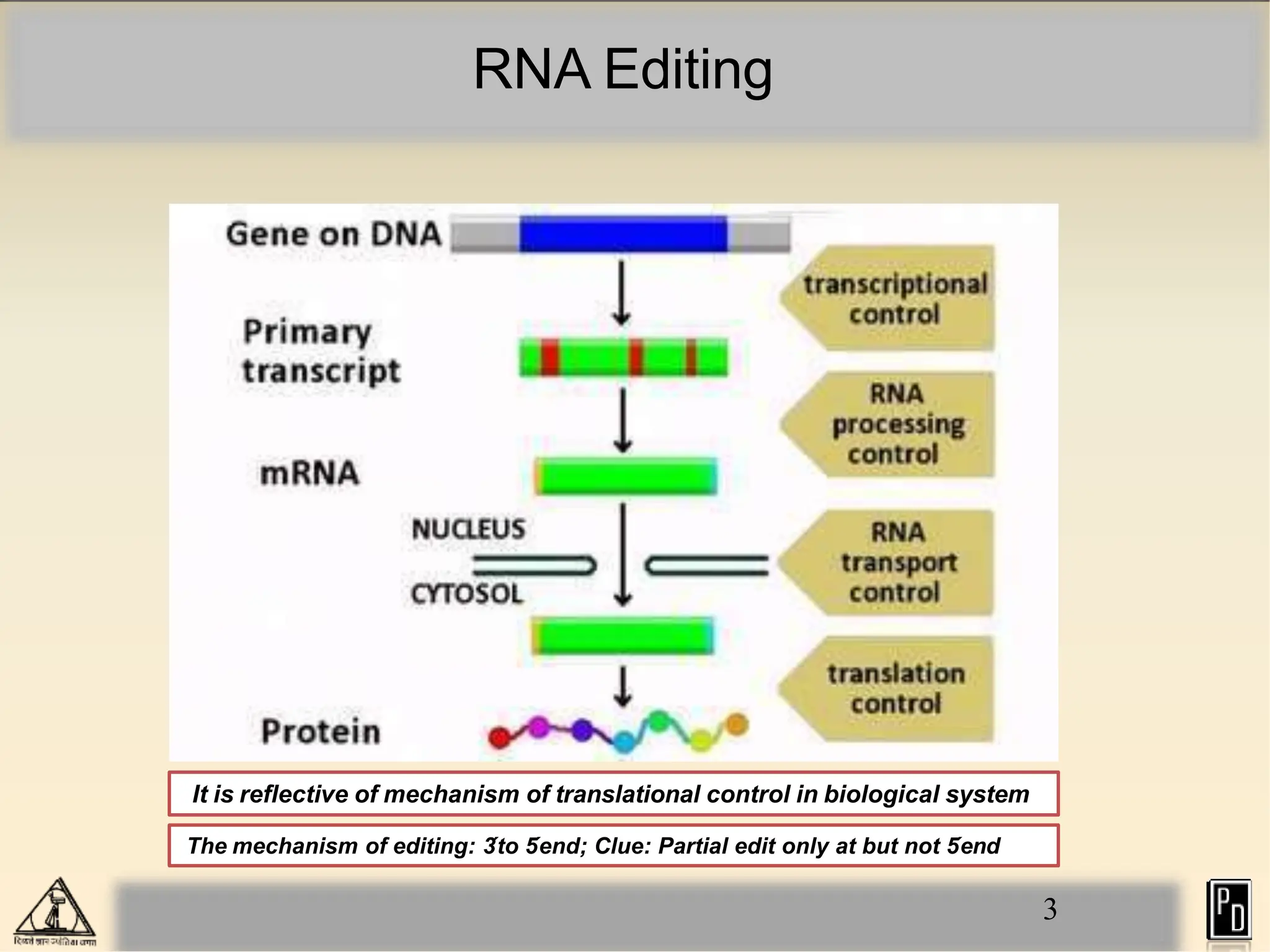
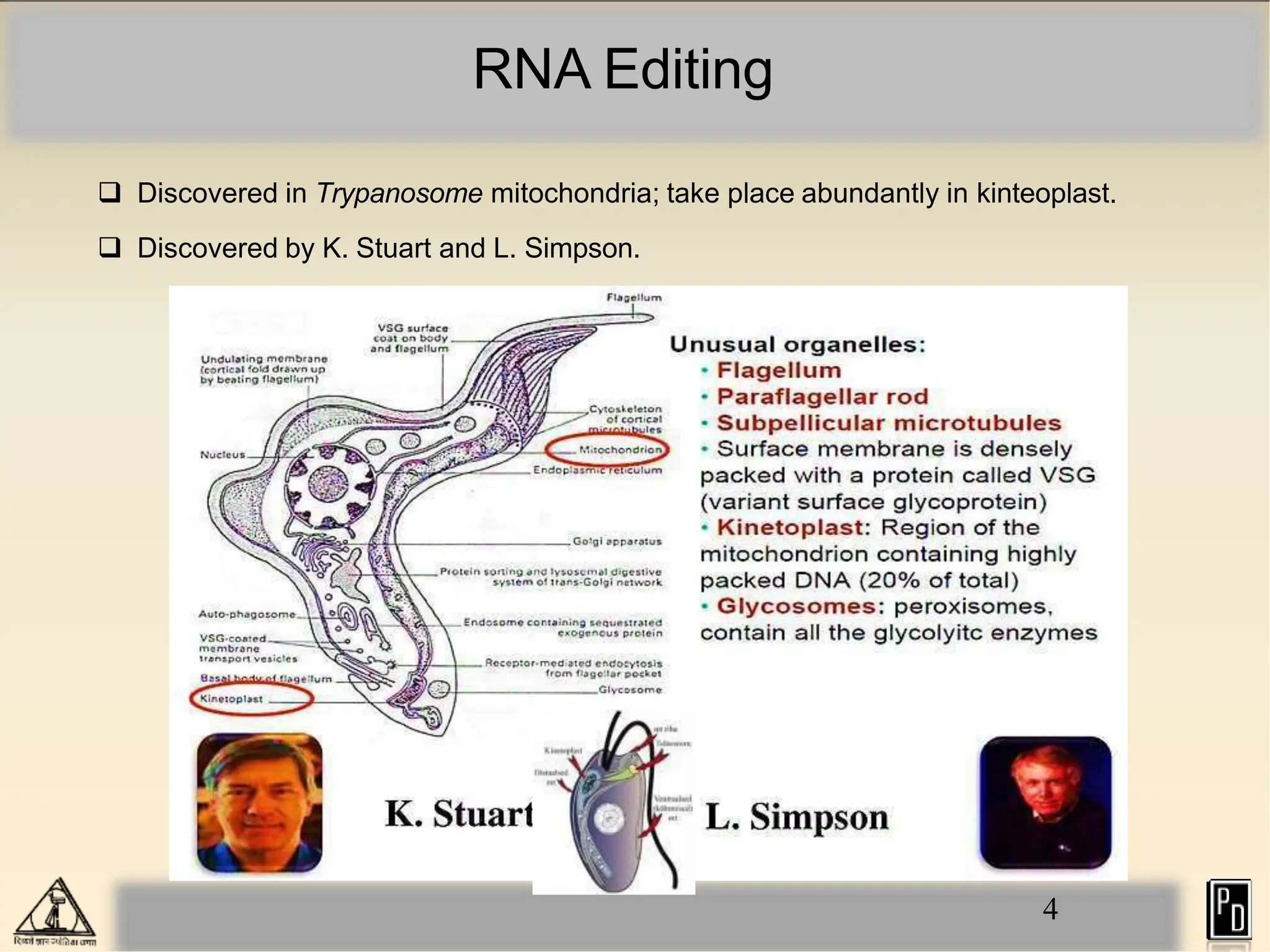
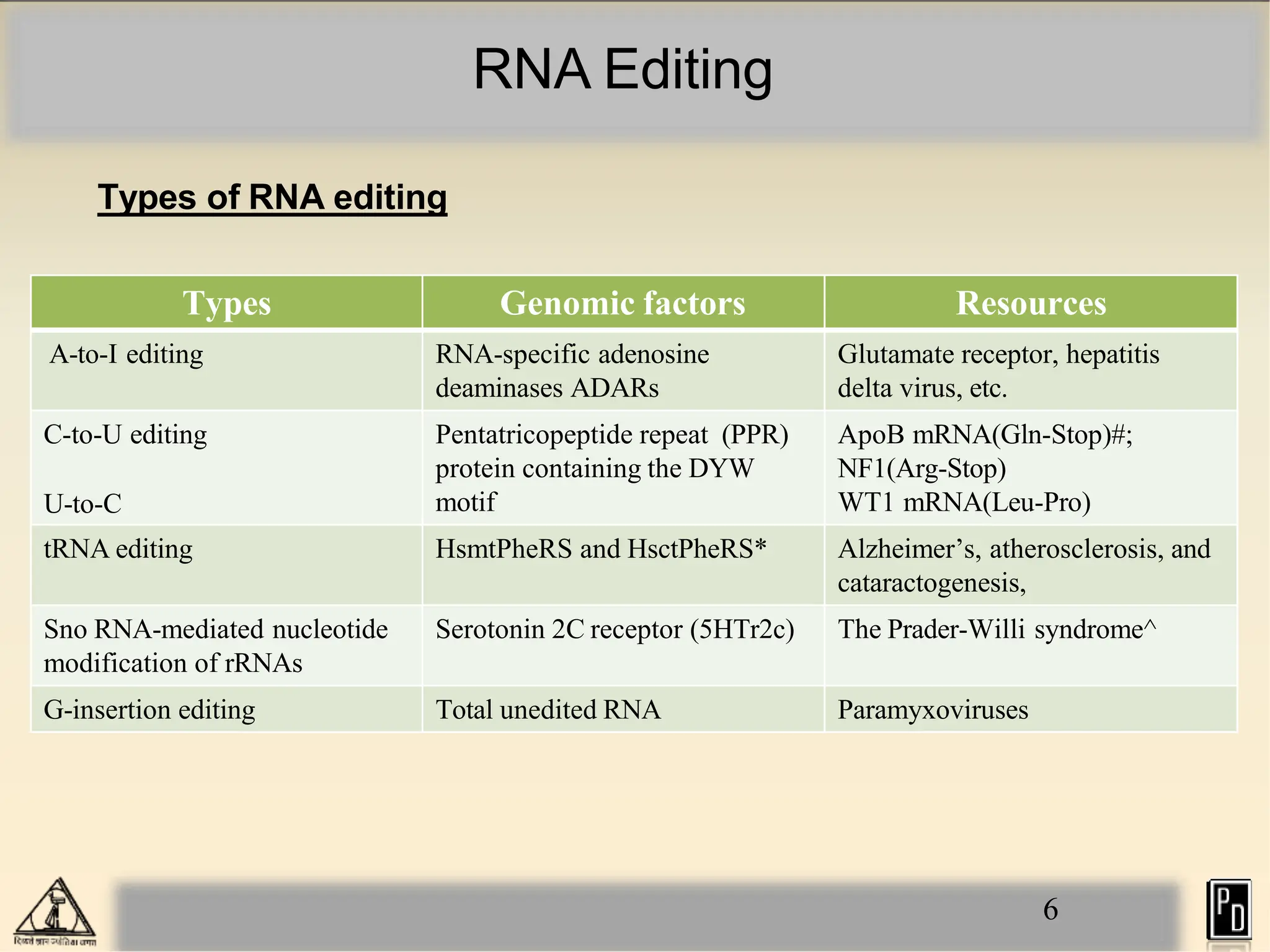
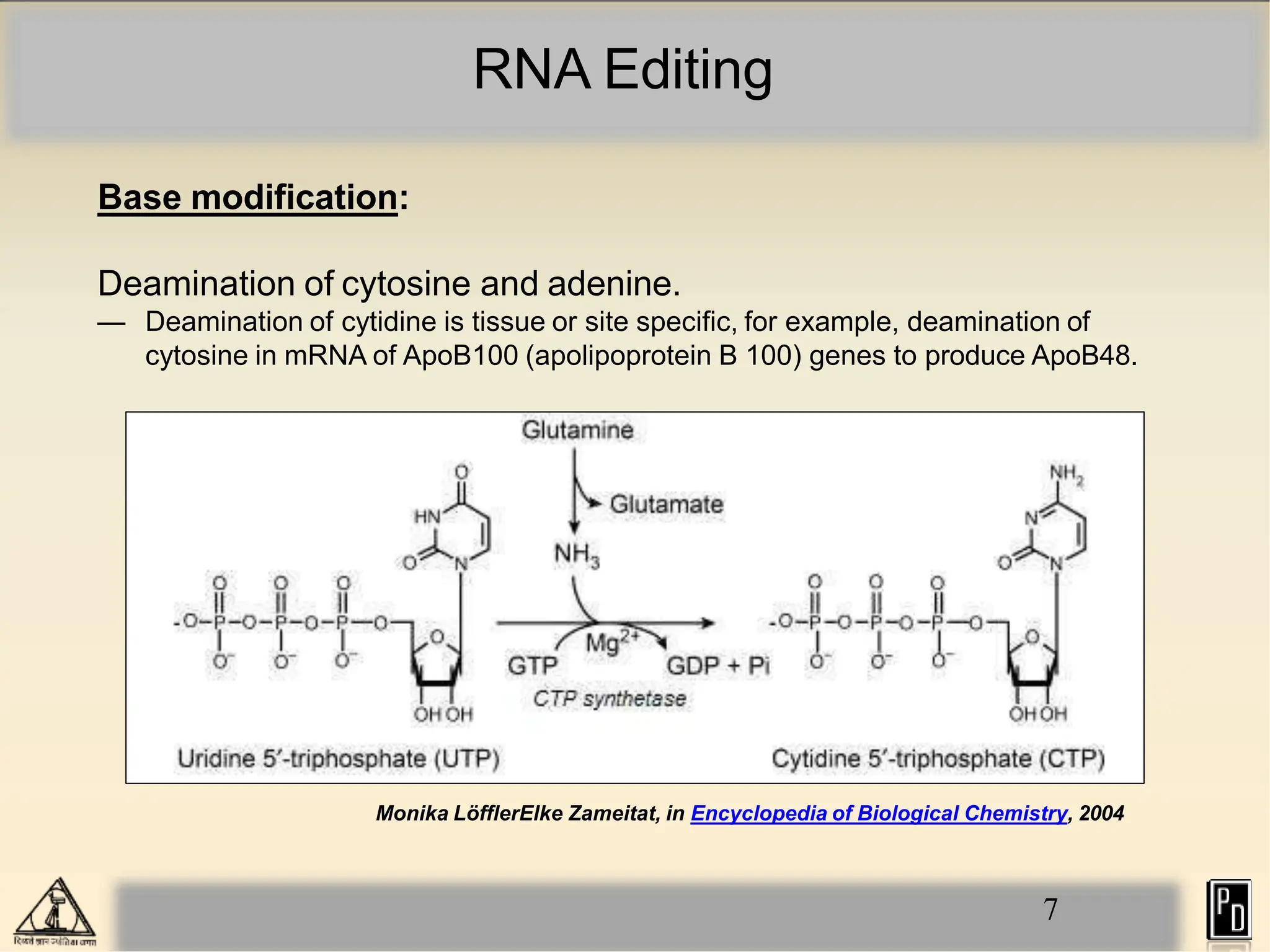
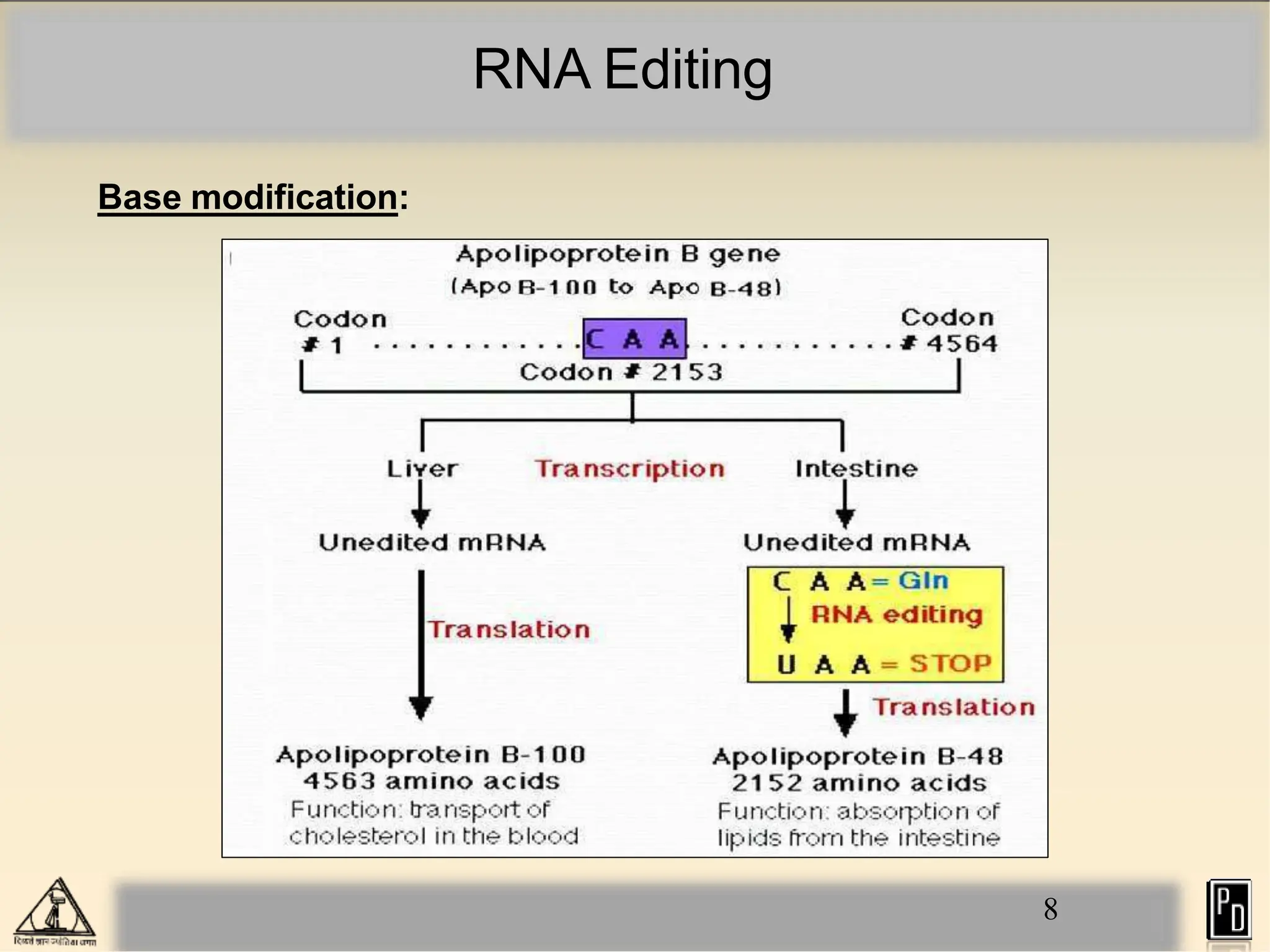
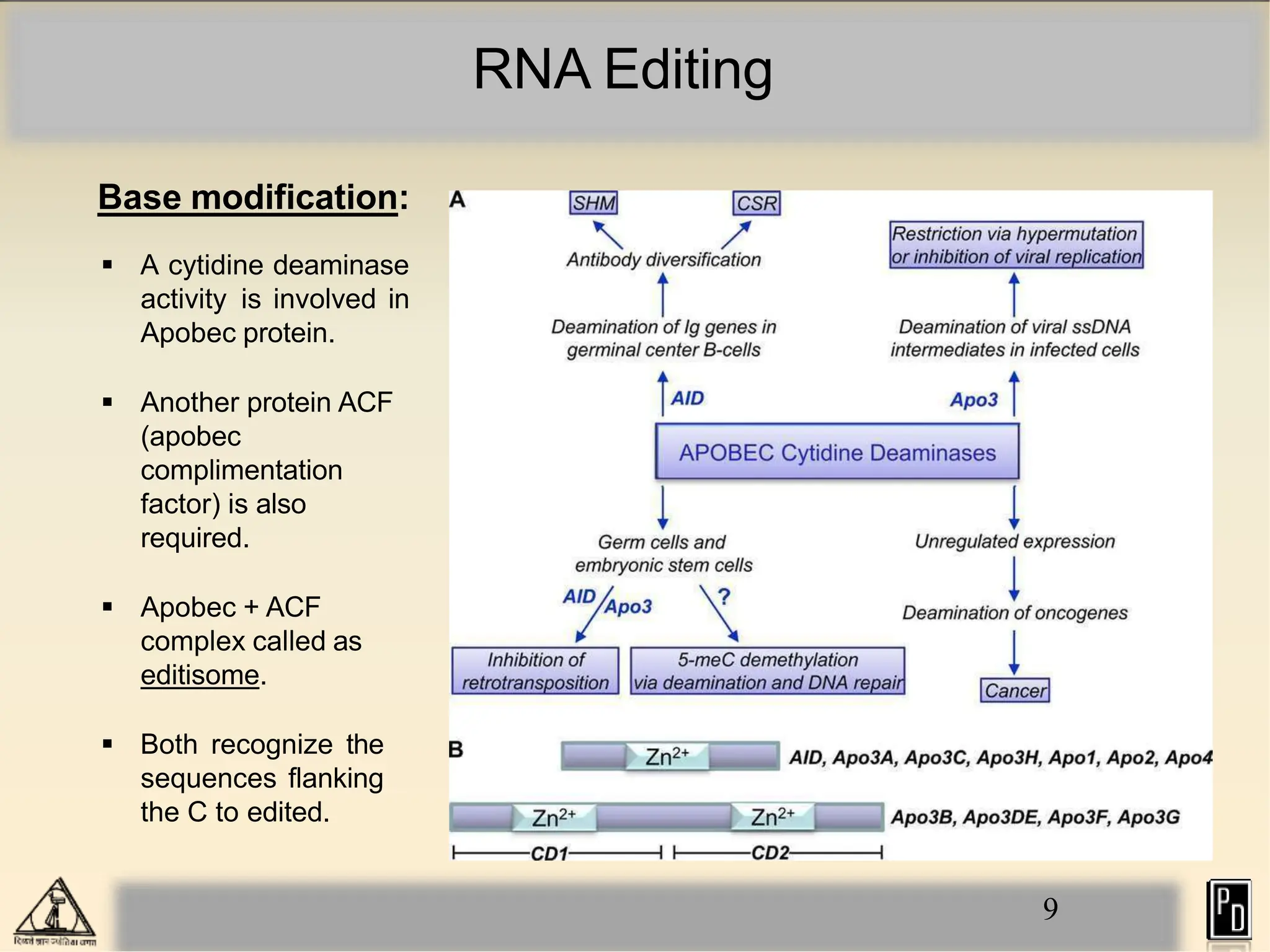
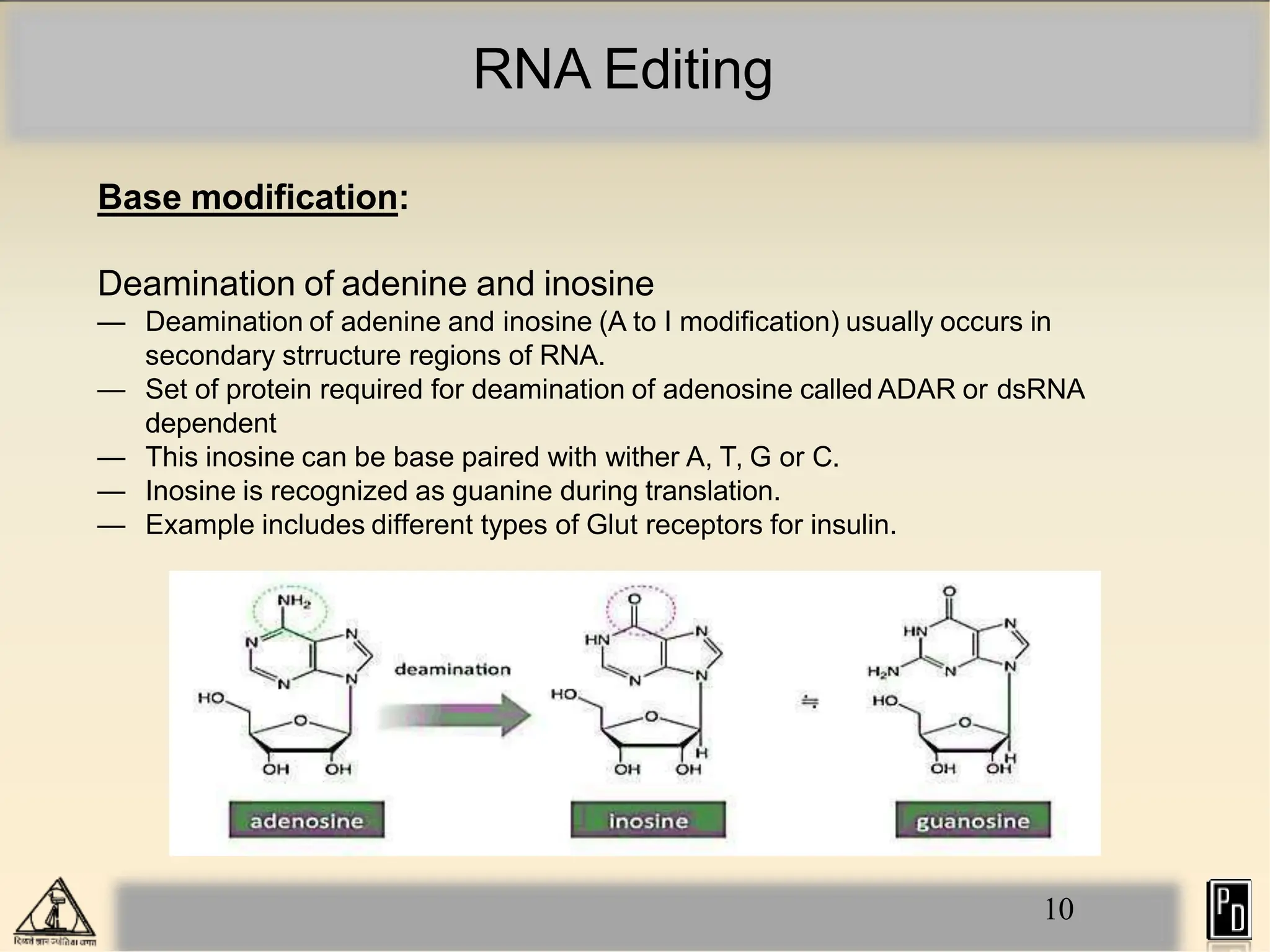
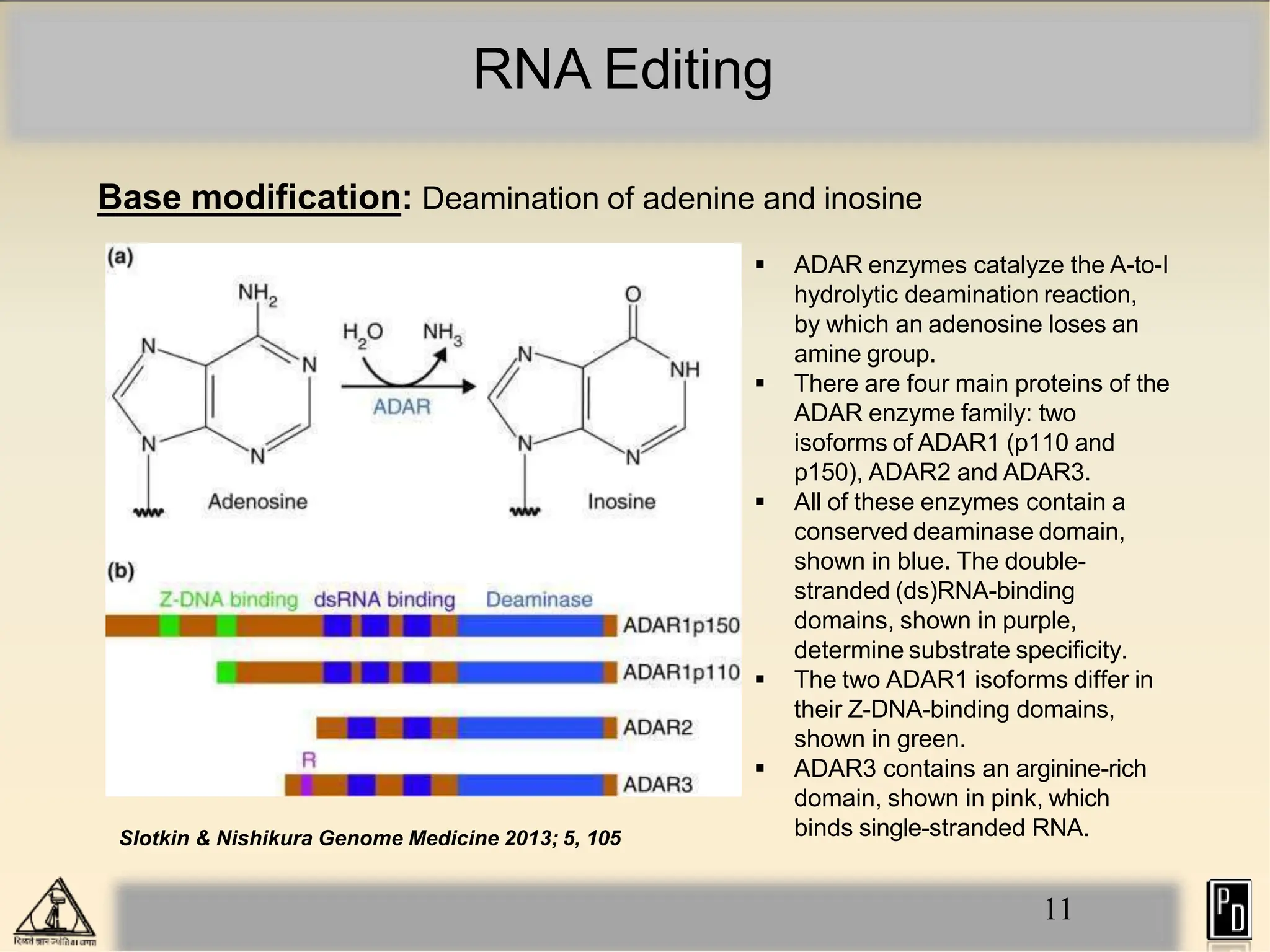
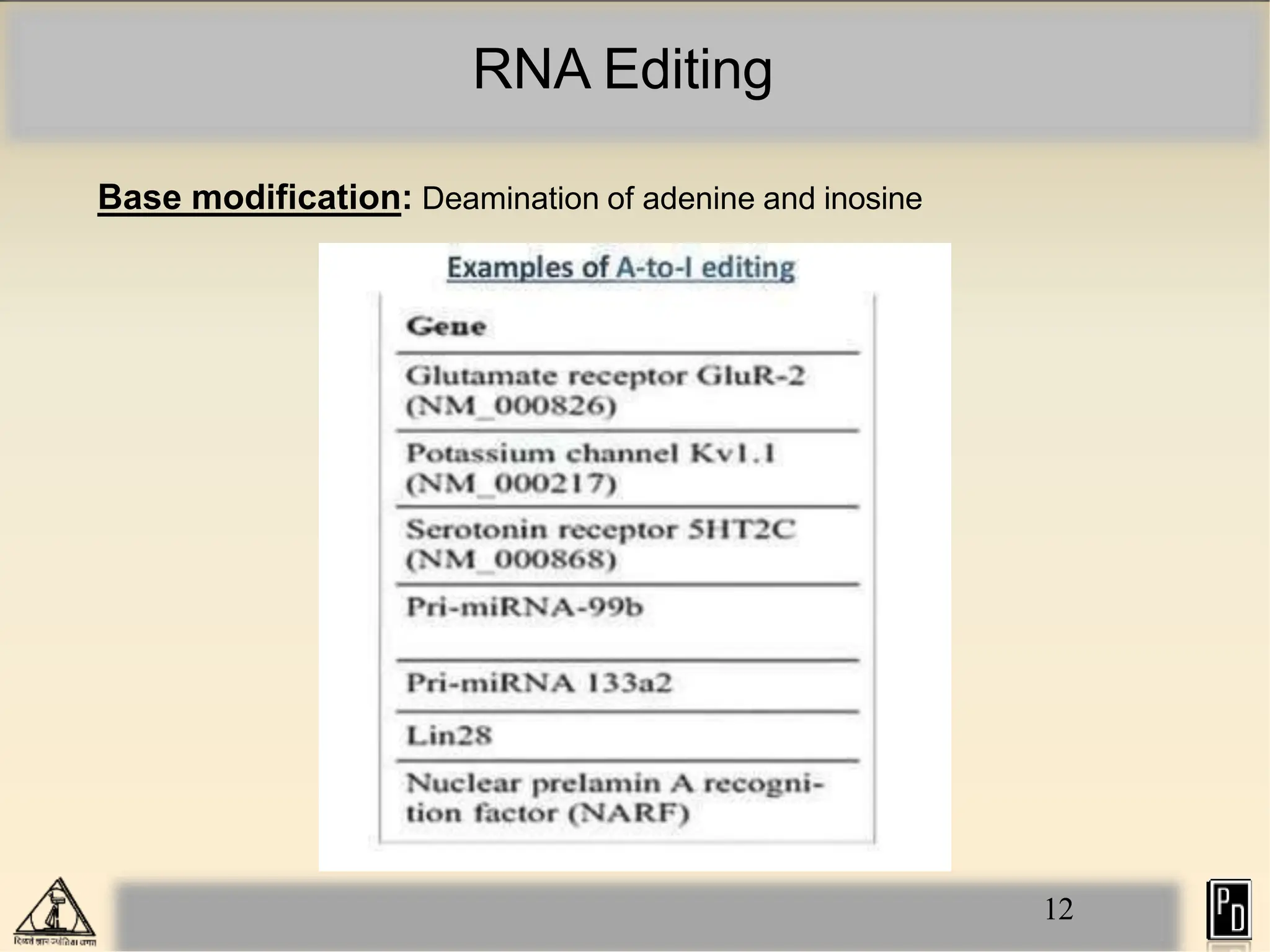
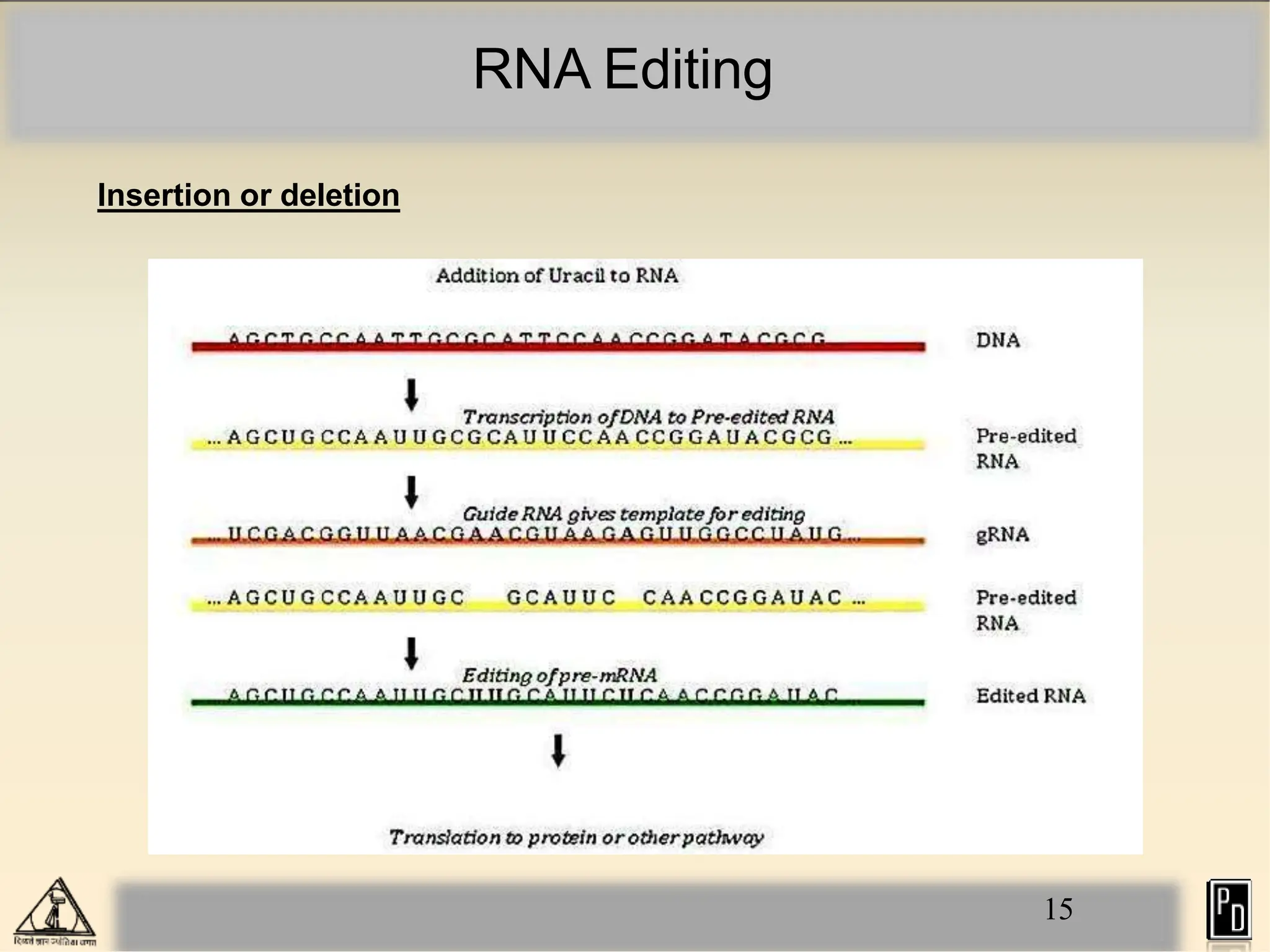
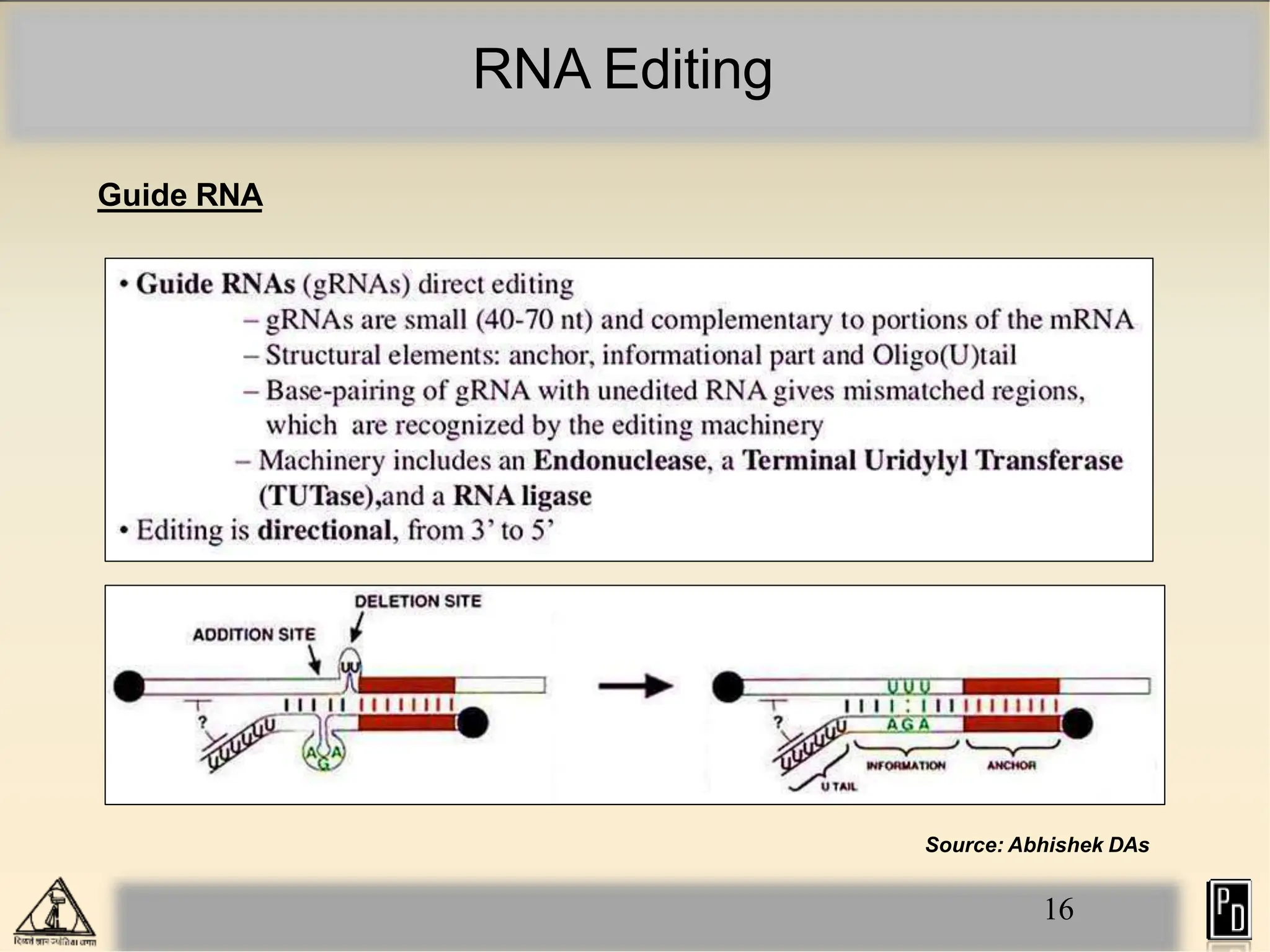
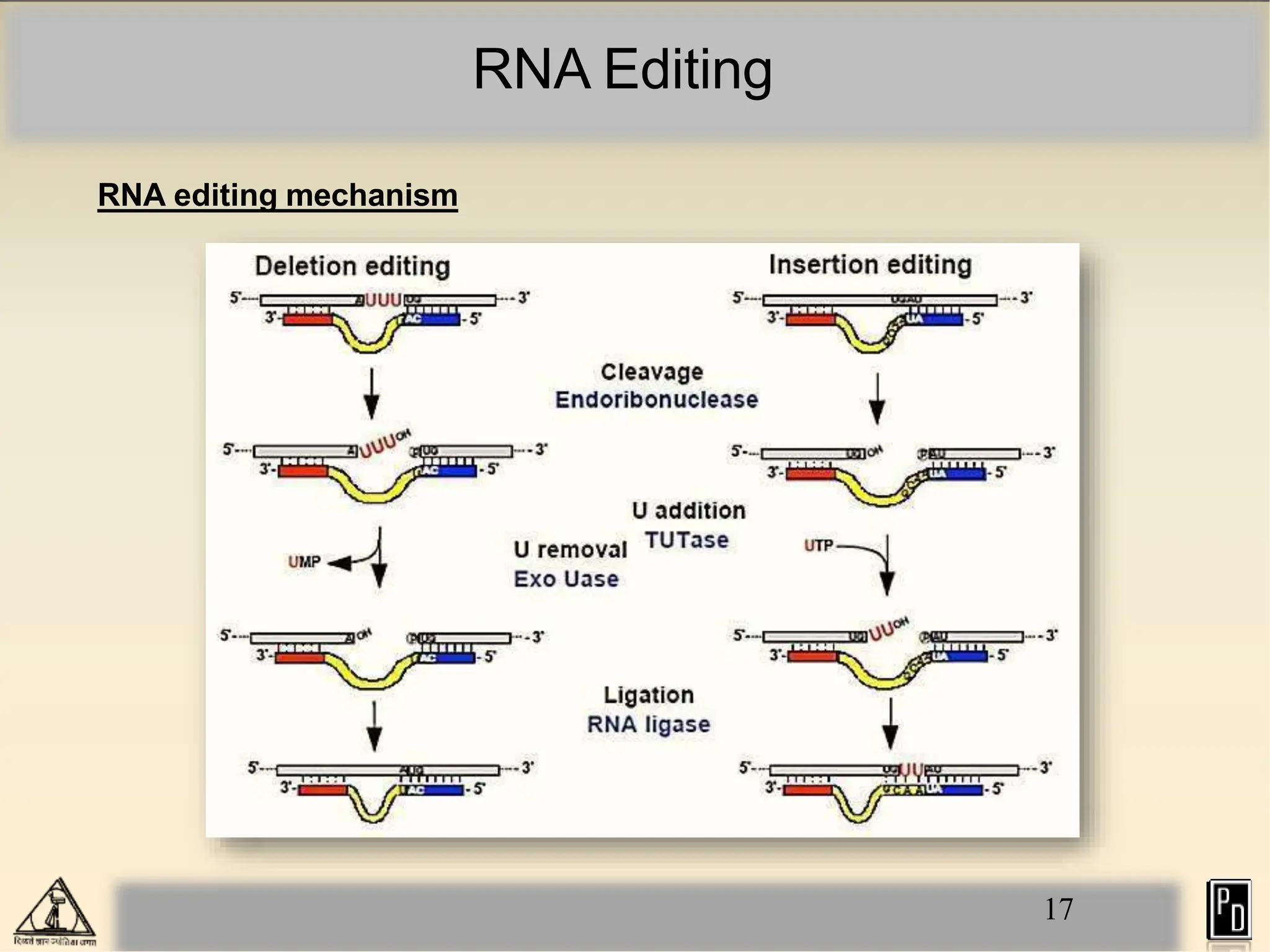
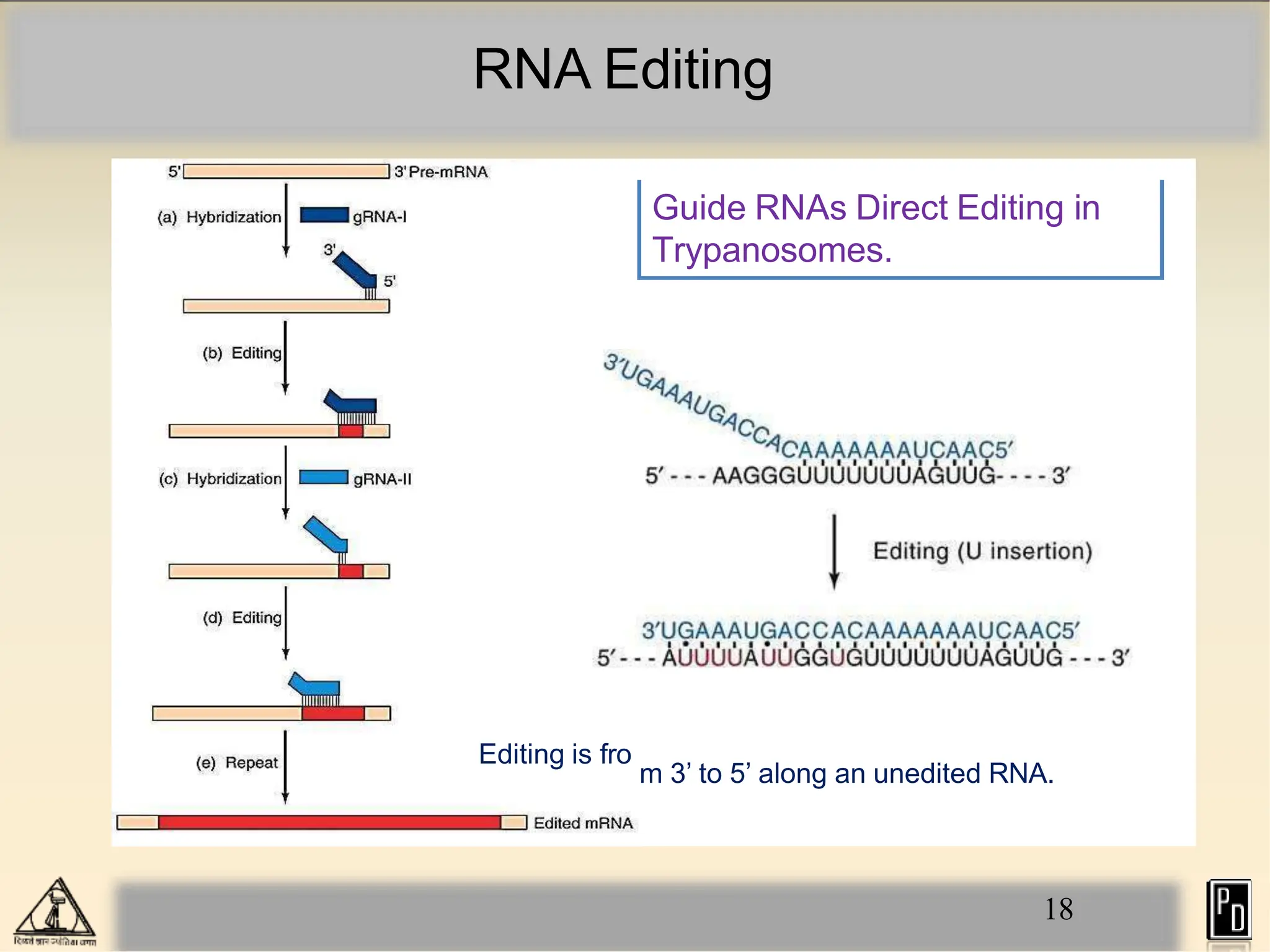
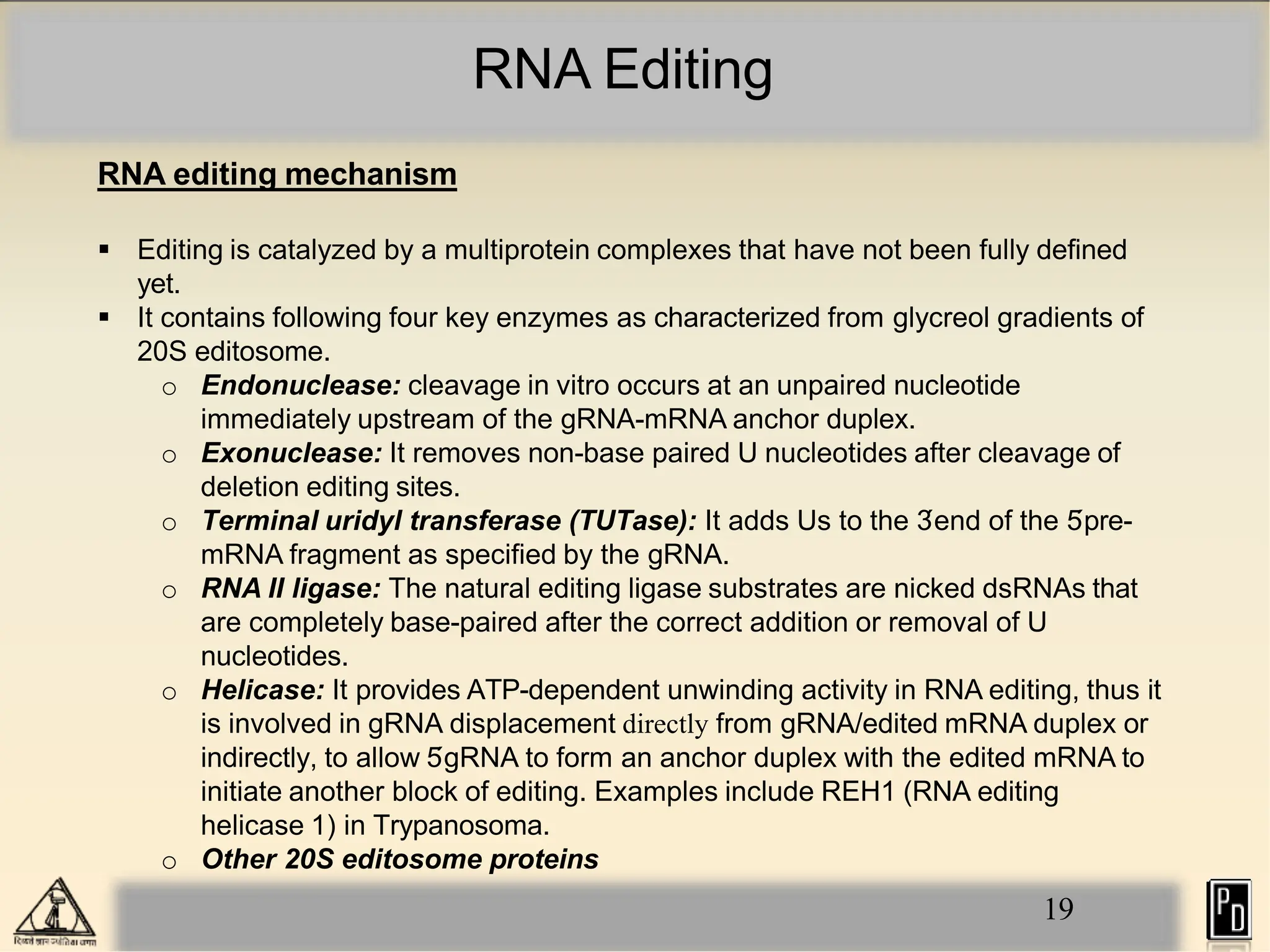
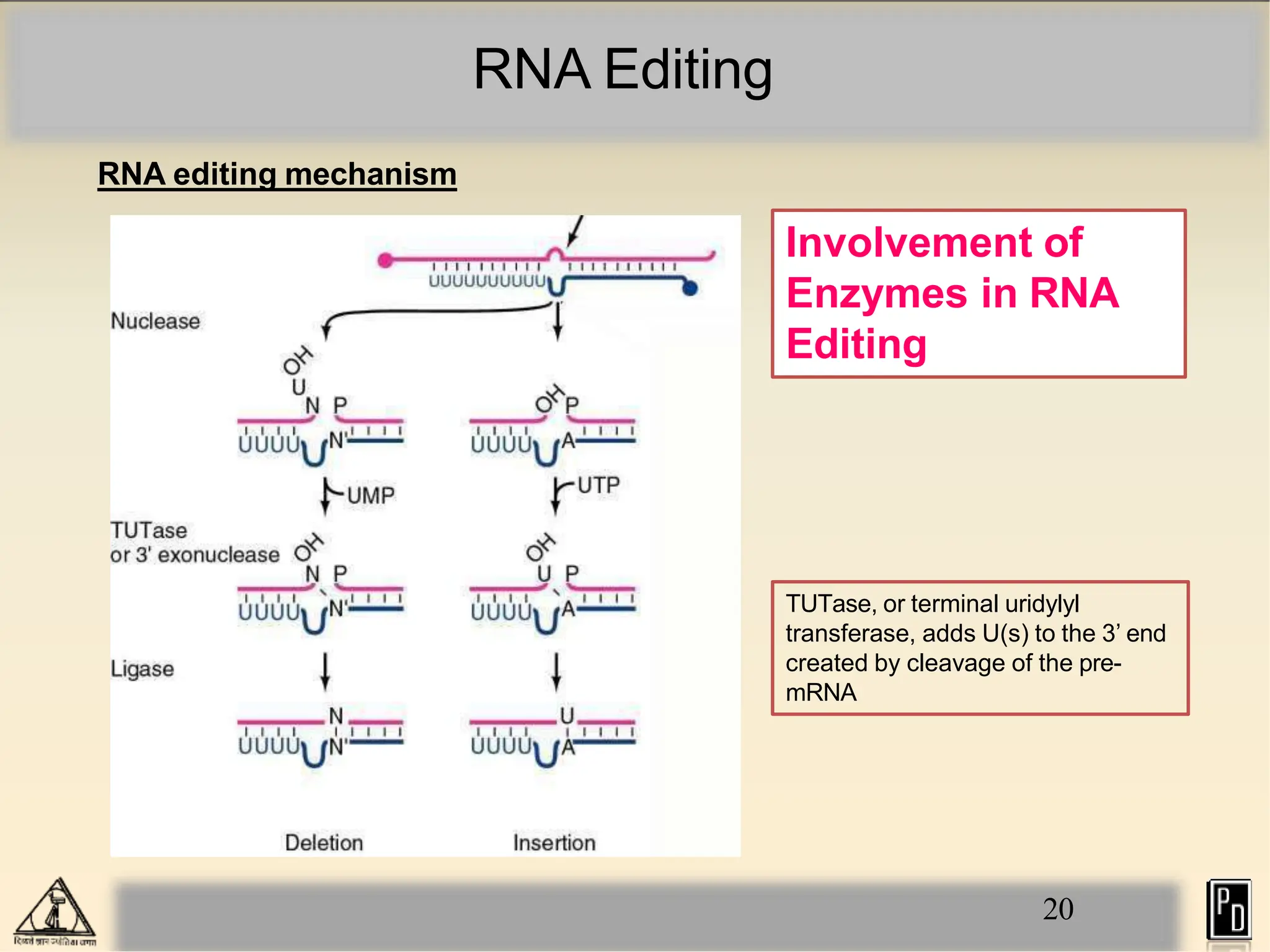
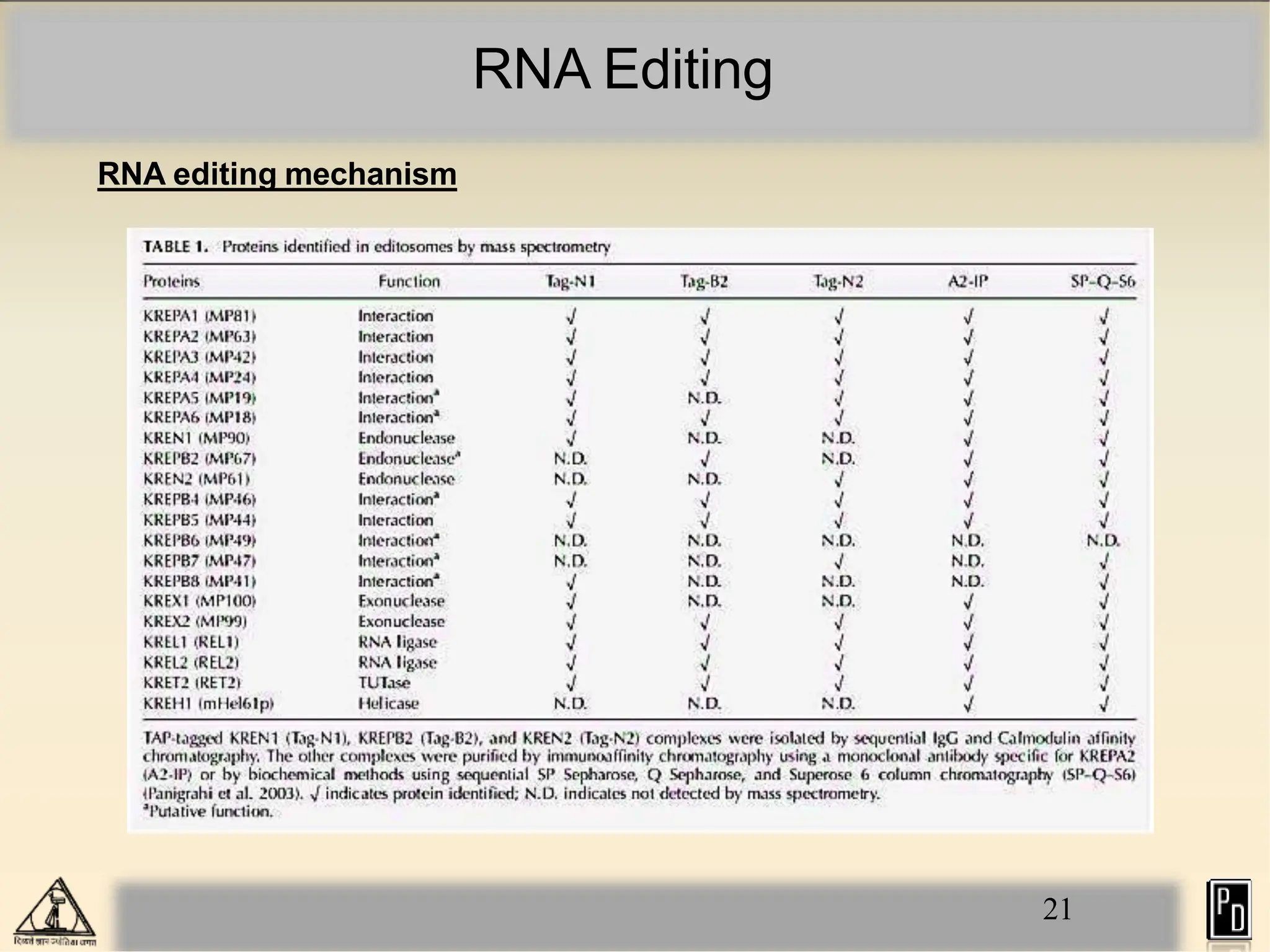
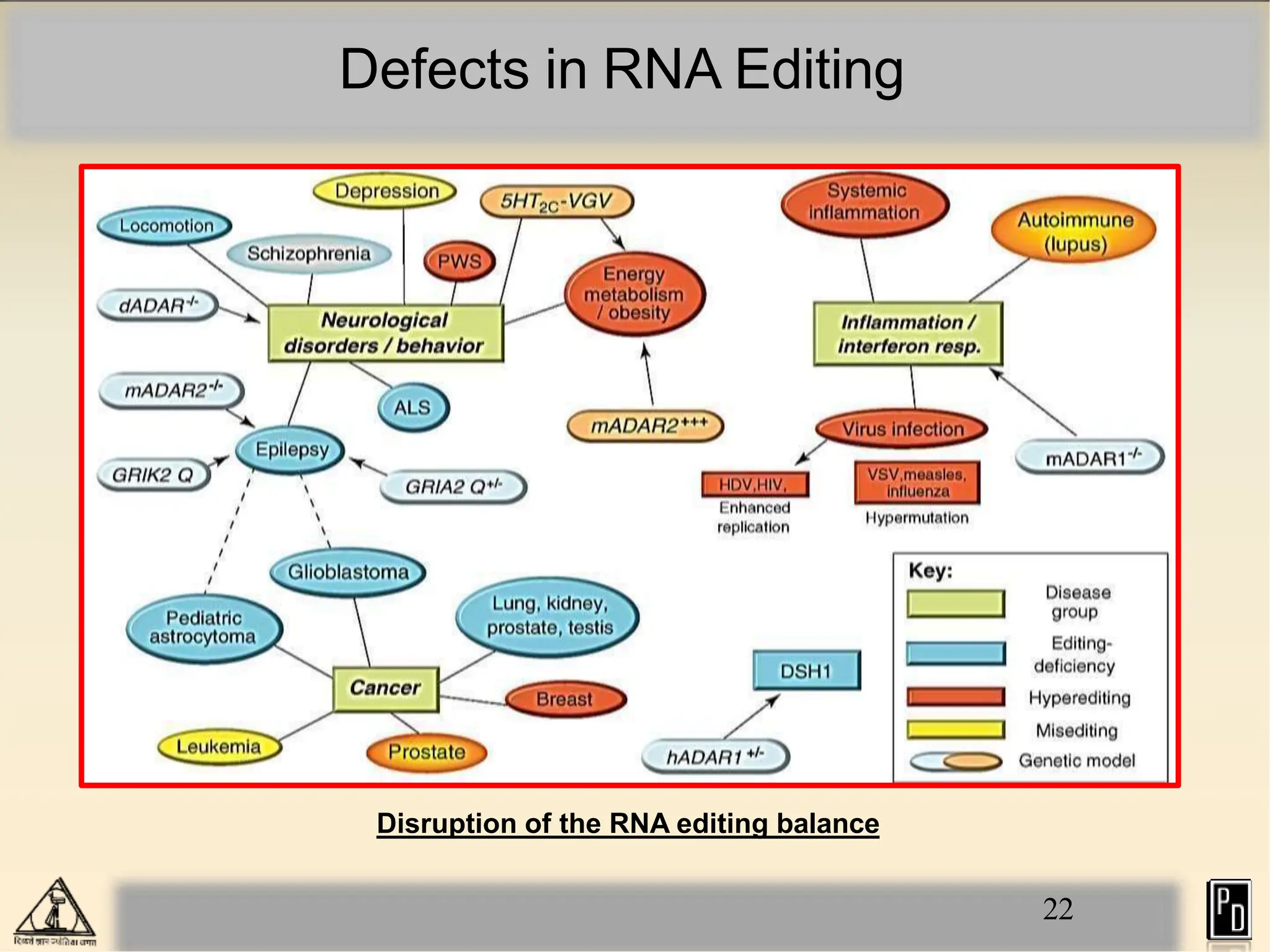
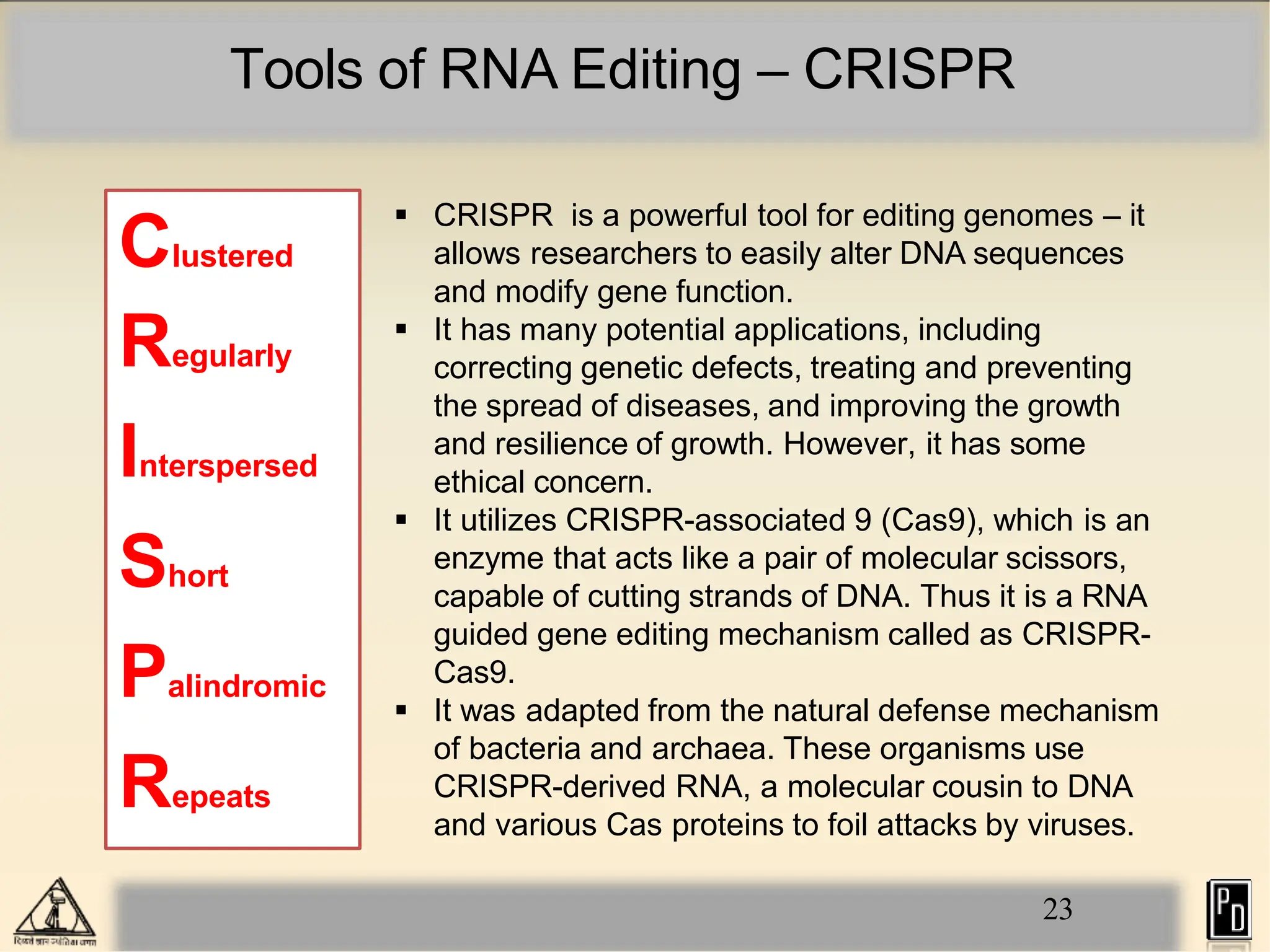
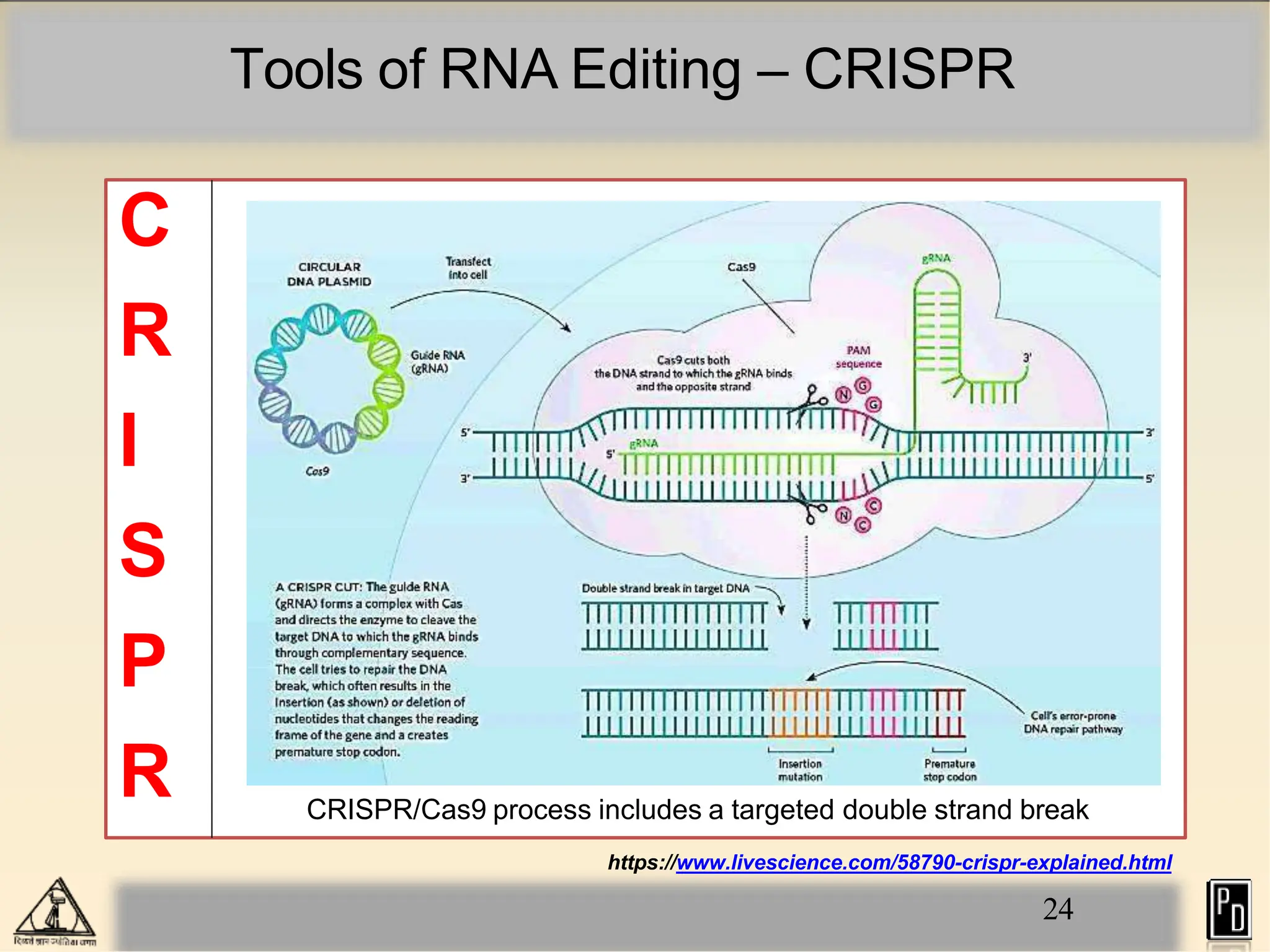
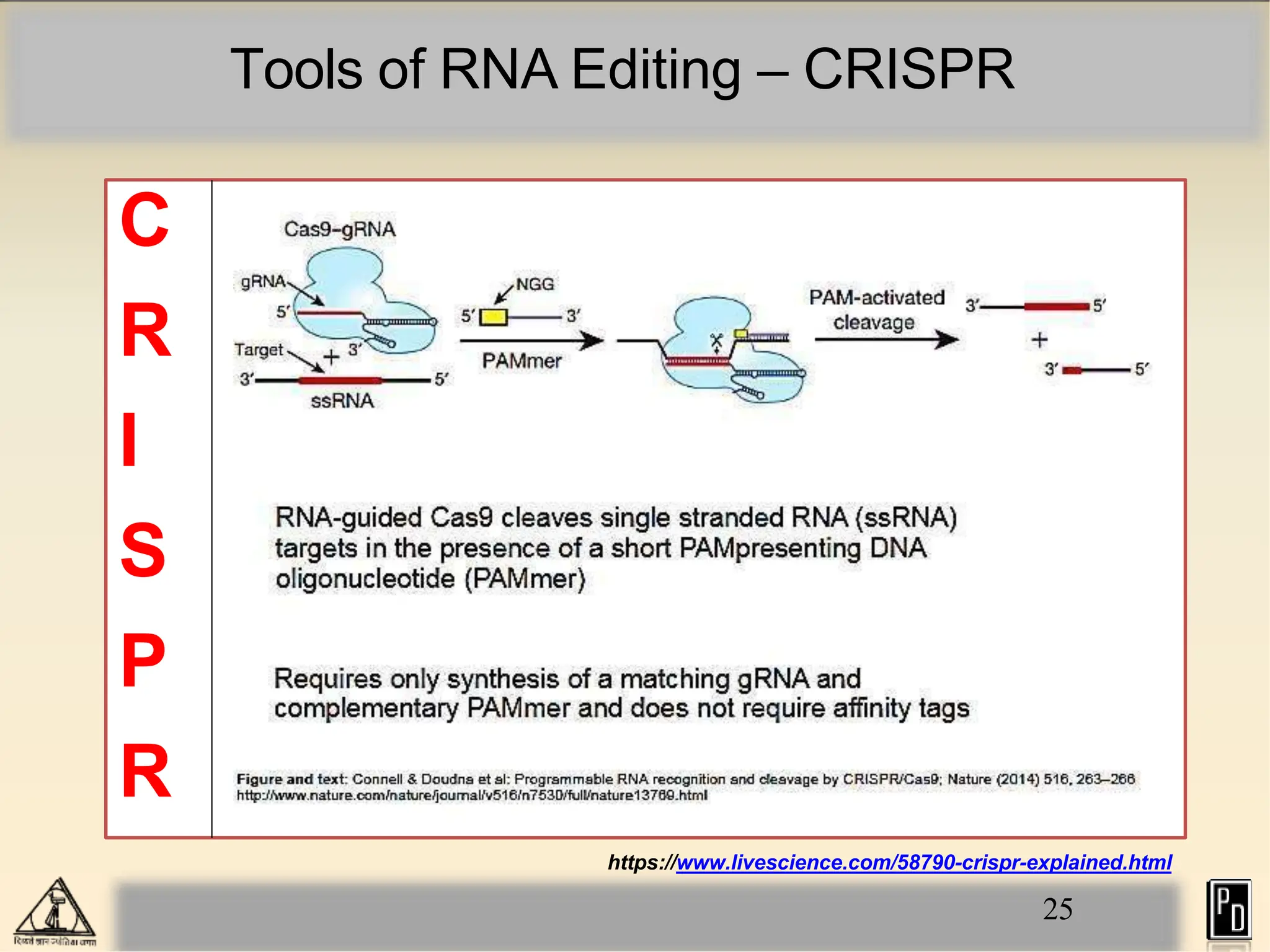
RNA editing is a post-transcriptional process that modifies RNA sequences without splicing. It was first discovered in trypanosomes and can occur in the nucleus, cytosol, mitochondria and plastids. There are two main types of RNA editing: base modification through deamination of adenine and cytosine, and insertion or deletion of uracil bases guided by trans-acting guide RNAs. RNA editing increases genetic diversity and is essential for regulating gene expression, though defects can cause disease. New tools like CRISPR also allow programmable RNA editing but raise ethical concerns.