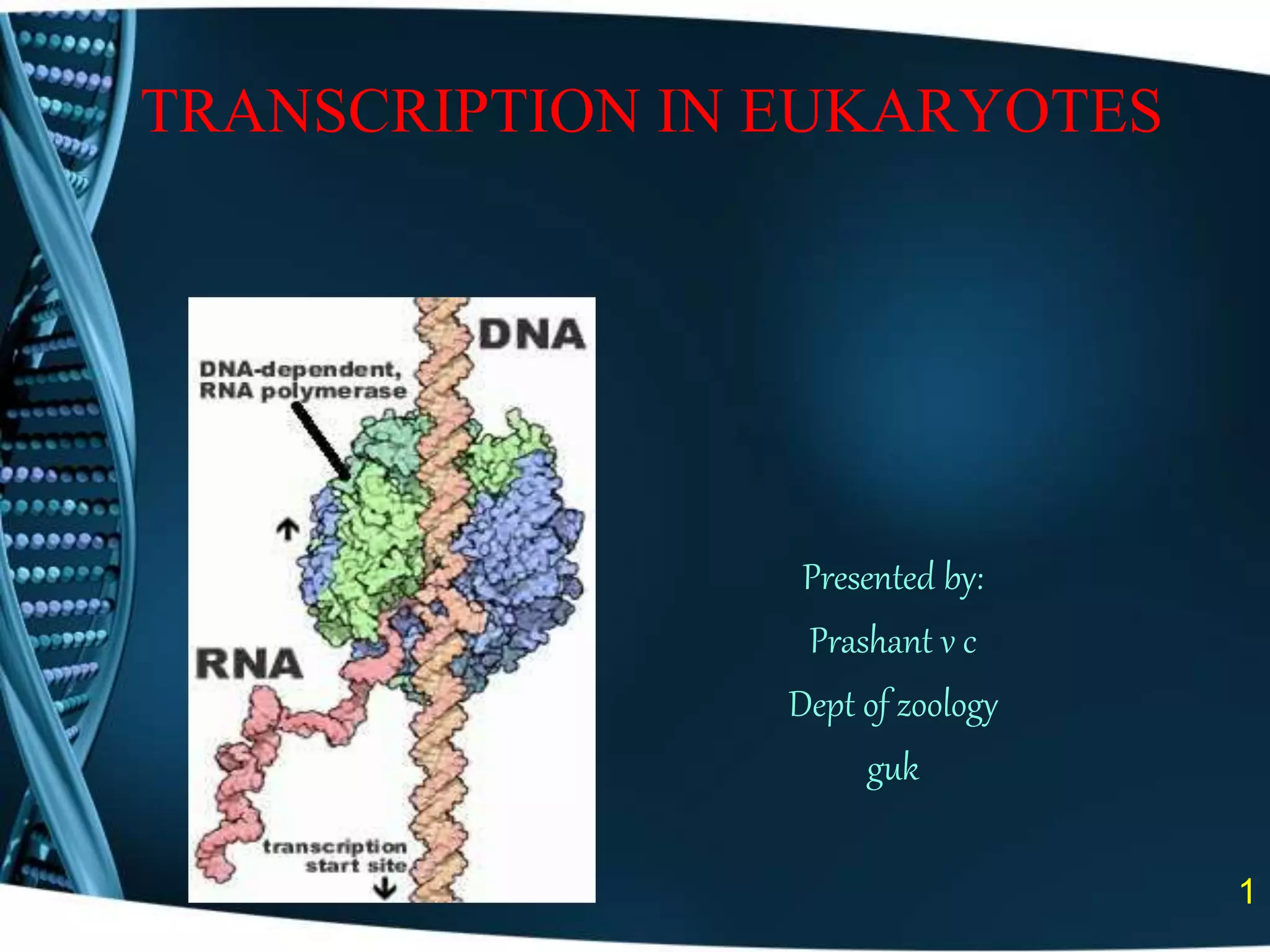
The document covers the process of transcription in eukaryotes, explaining the roles of different types of RNA, the structure of genes, and the mechanisms of transcription including initiation, elongation, and termination. It highlights the importance of transcription factors, chromatin modifying proteins, and RNA splicing in regulating gene expression. The document contrasts eukaryotic and prokaryotic transcription and outlines the involvement of RNA polymerases in synthesizing various RNA types.