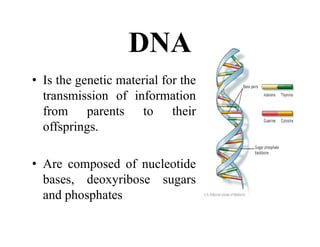
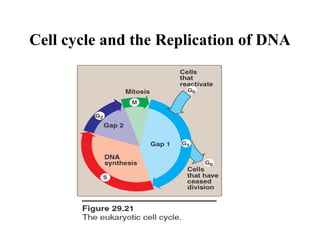
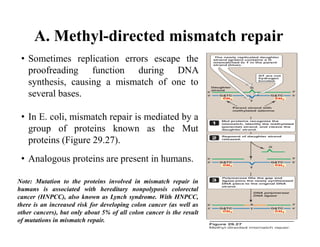
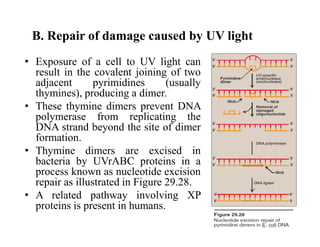
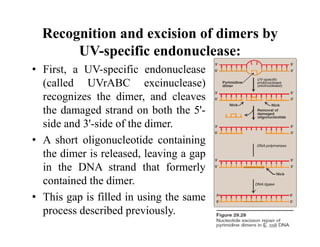
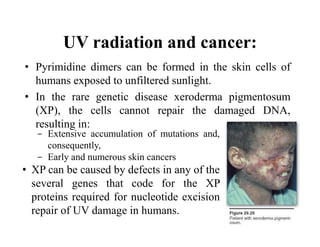
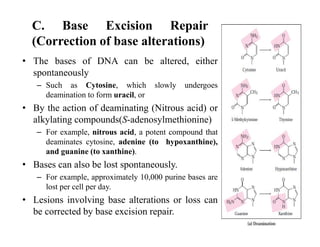
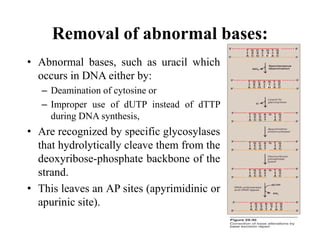
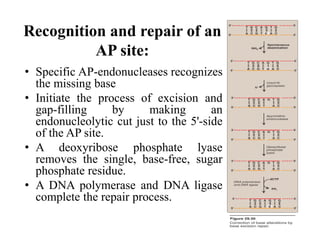
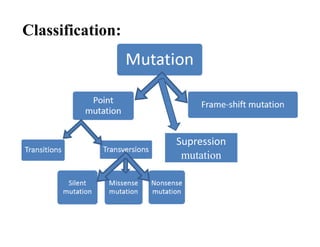
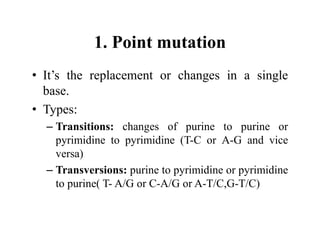
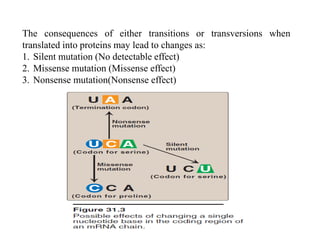
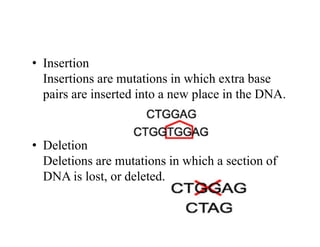
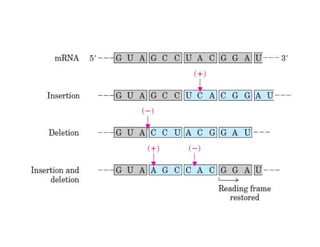
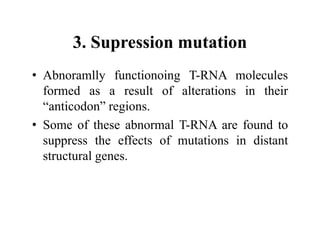
DNA contains the genetic information that is passed from parents to offspring. It is composed of nucleotide bases, sugars, and phosphates. DNA can be damaged by environmental factors or errors during replication. Cells have multiple repair systems to fix DNA damage through processes like base excision repair, nucleotide excision repair, and mismatch repair. Unrepaired damage may lead to mutations, which are heritable changes in DNA sequence that can cause disease if they affect important genes.