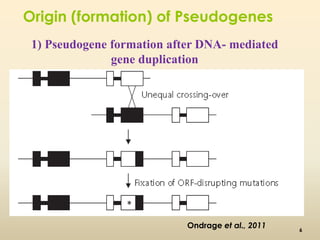
This document discusses pseudogenes, which are dysfunctional copies of genes that have lost protein-coding ability. It covers the origin and formation of pseudogenes through DNA or RNA duplication, and describes different types like processed and unprocessed pseudogenes. The document also discusses various methods for identifying and detecting pseudogenes, databases of pseudogenes, and studies that have characterized pseudogenes in organisms like rice and Solanum plants. Finally, it explores the potential functions and utilities of pseudogenes, including their use in evolutionary studies, providing information about gene expression, and acting as competing endogenous RNAs.