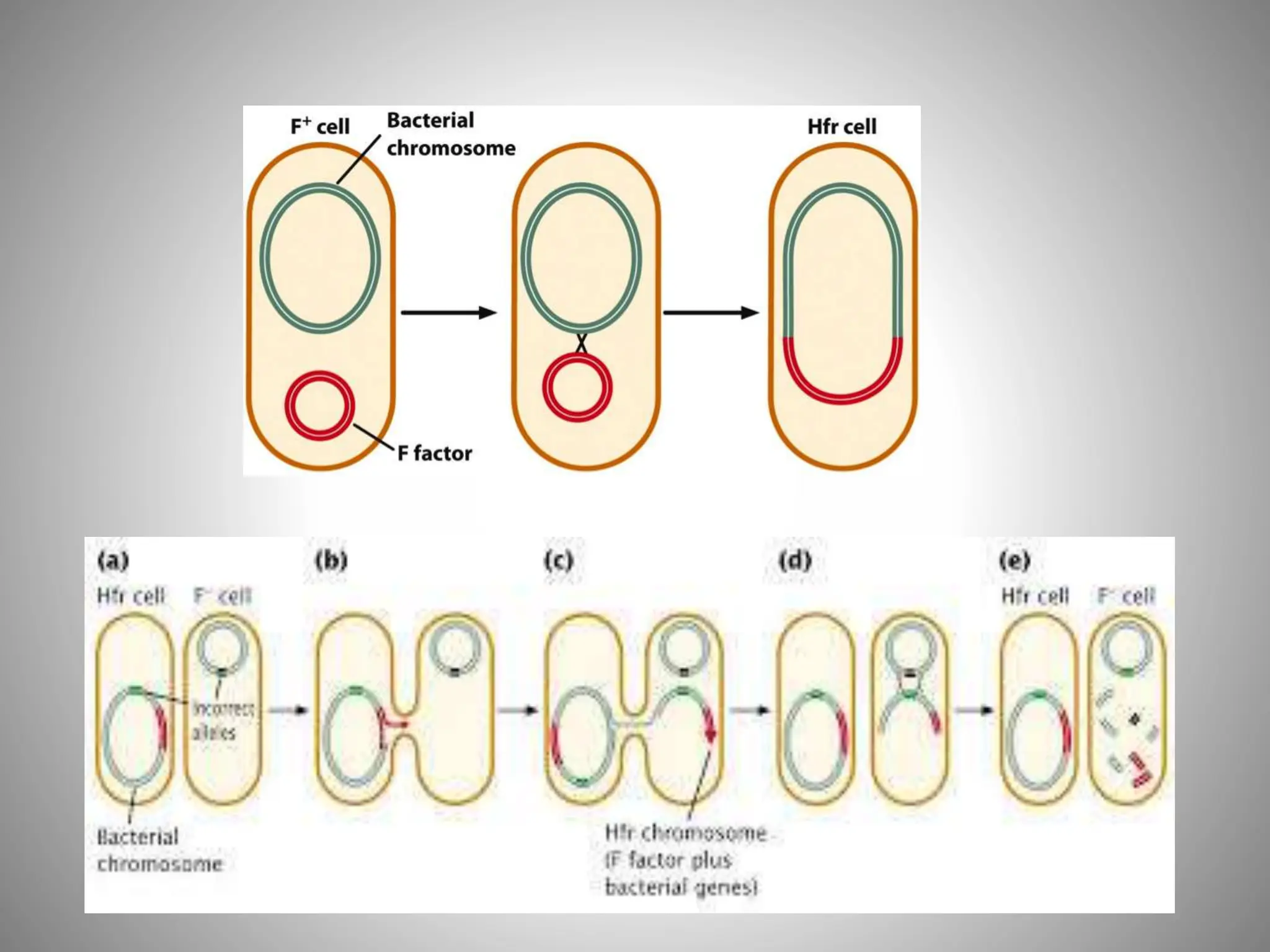
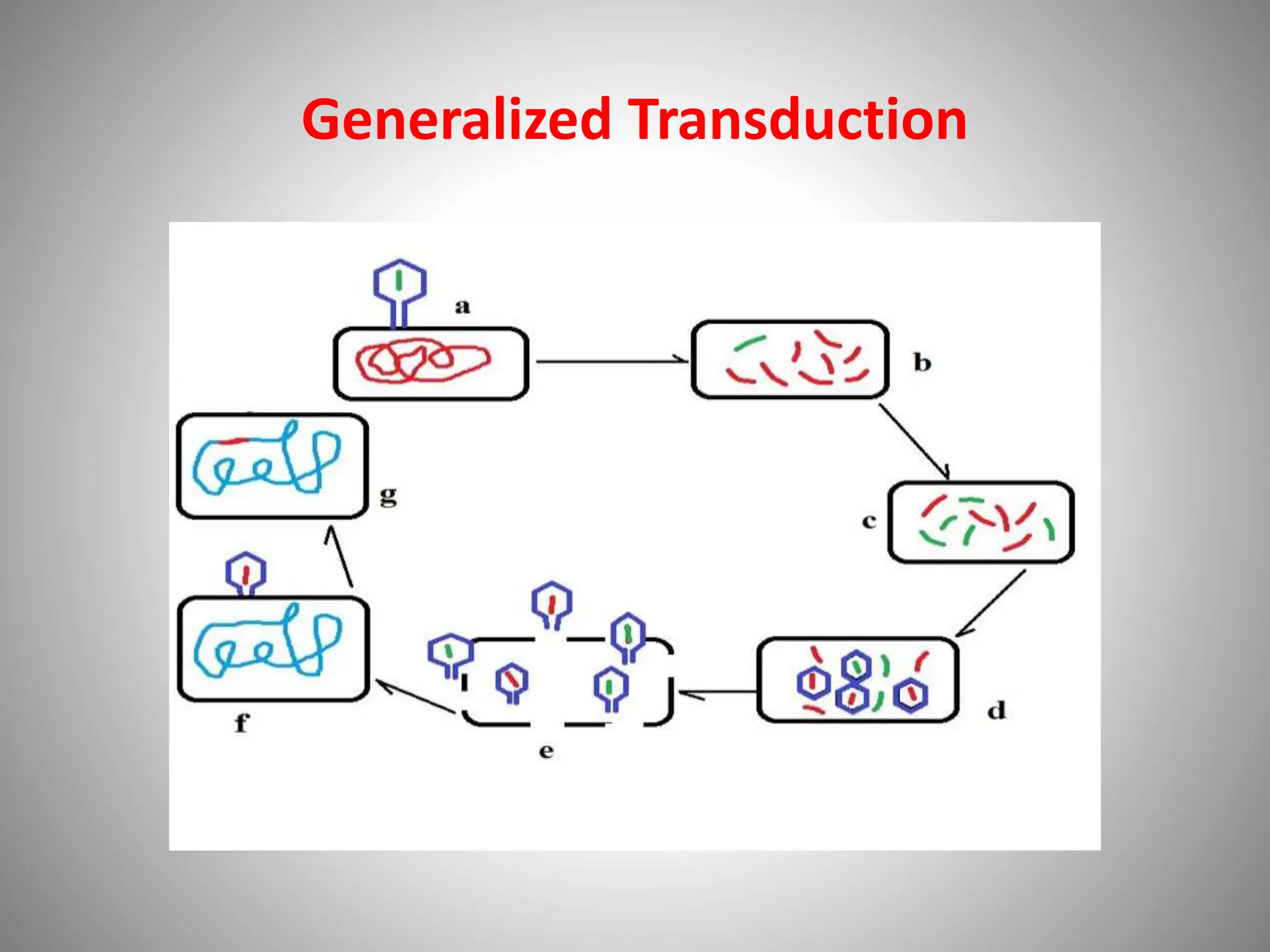
The document discusses microbial genetic recombinations, highlighting bacterial recombination methods such as conjugation, transduction, and transformation. It elaborates on the mechanics of each process, including the role of plasmids and transposons in gene transfer and genetic variability among microorganisms. Additionally, it outlines the discoveries and principles associated with each form of recombination, emphasizing their significance in microbial genetics.