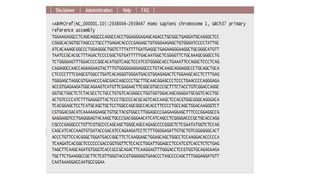
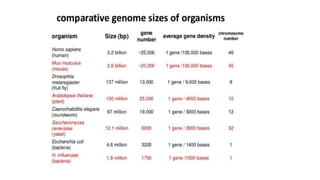
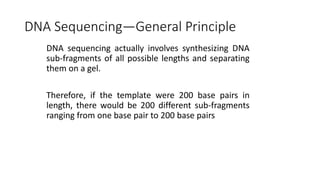
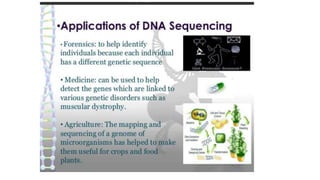
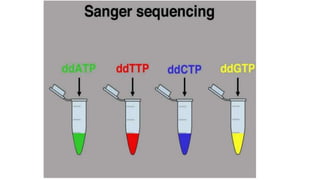
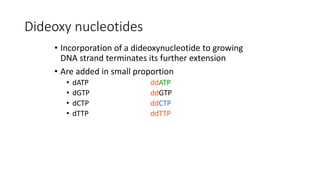
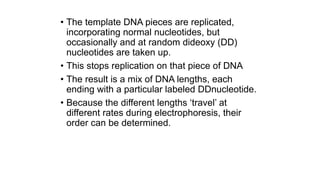
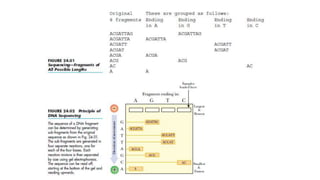
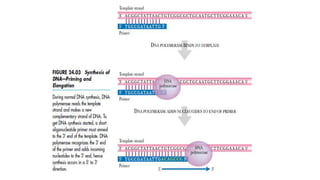
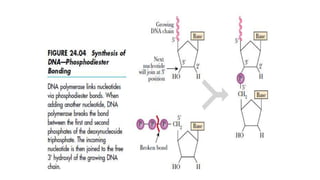
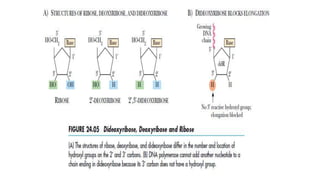
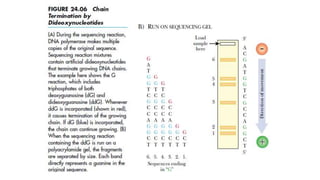
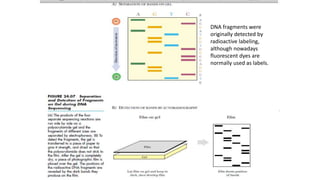
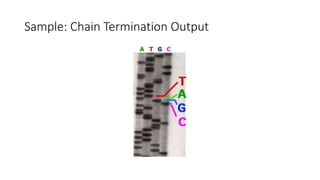
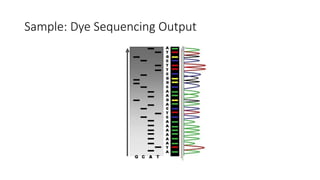
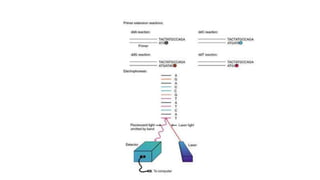
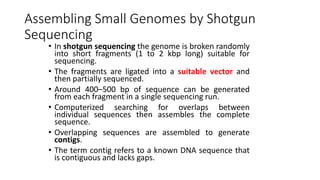
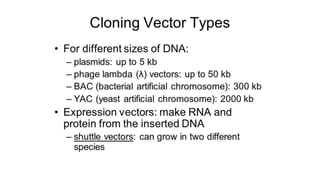
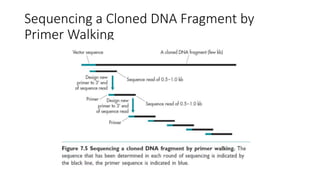
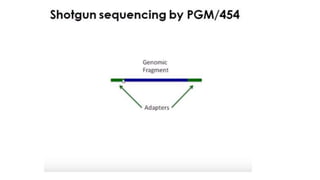
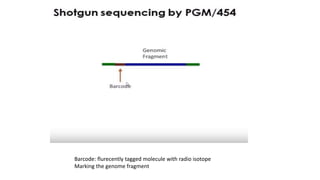
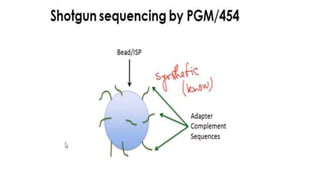
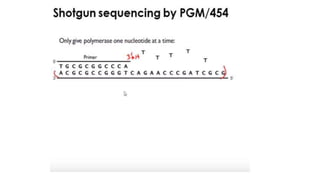
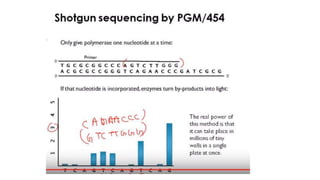
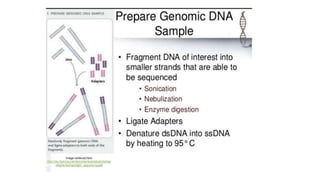
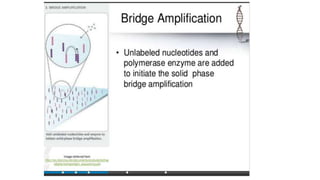
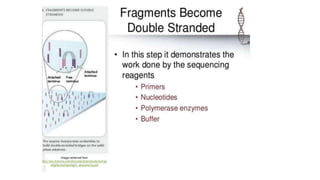
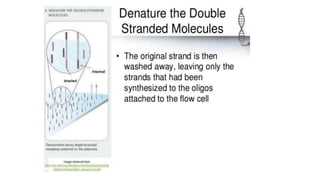
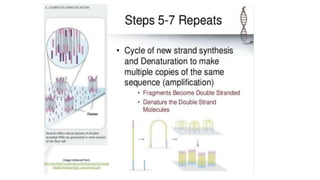
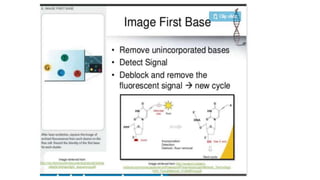
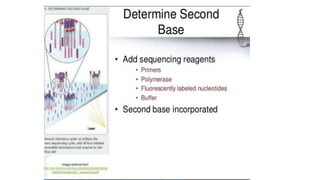
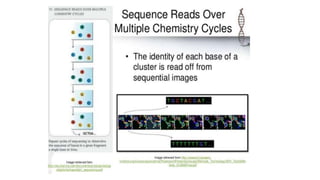
DNA sequencing involves determining the order of nucleotides in a DNA molecule. There are several methods for DNA sequencing, including Sanger sequencing using chain termination with dideoxynucleotides, Maxam-Gilbert chemical sequencing, shotgun sequencing by fragmenting DNA into random pieces, and newer next-generation sequencing technologies like Illumina sequencing and Ion Torrent sequencing that are faster and cheaper. Understanding DNA sequences can provide insight into genetic conditions and diseases and has applications in medicine, agriculture, and forensics.