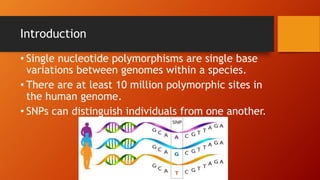
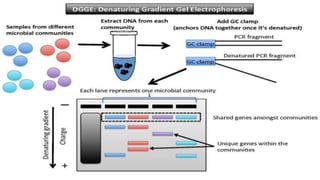
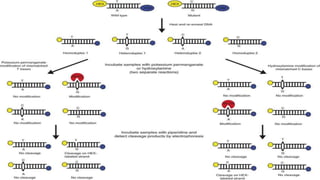
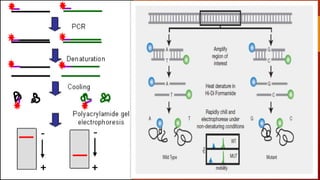
The document discusses single nucleotide polymorphisms (SNPs), which are variations in DNA that can distinguish individuals and are prevalent in the human genome. It outlines various methods for SNP detection, including both global and targeted discovery techniques, and details specific technologies such as denaturing gradient gel electrophoresis and Muts protein-binding assays. Additionally, the applications of SNP detection in fields like agriculture and pharmacogenetics are highlighted.