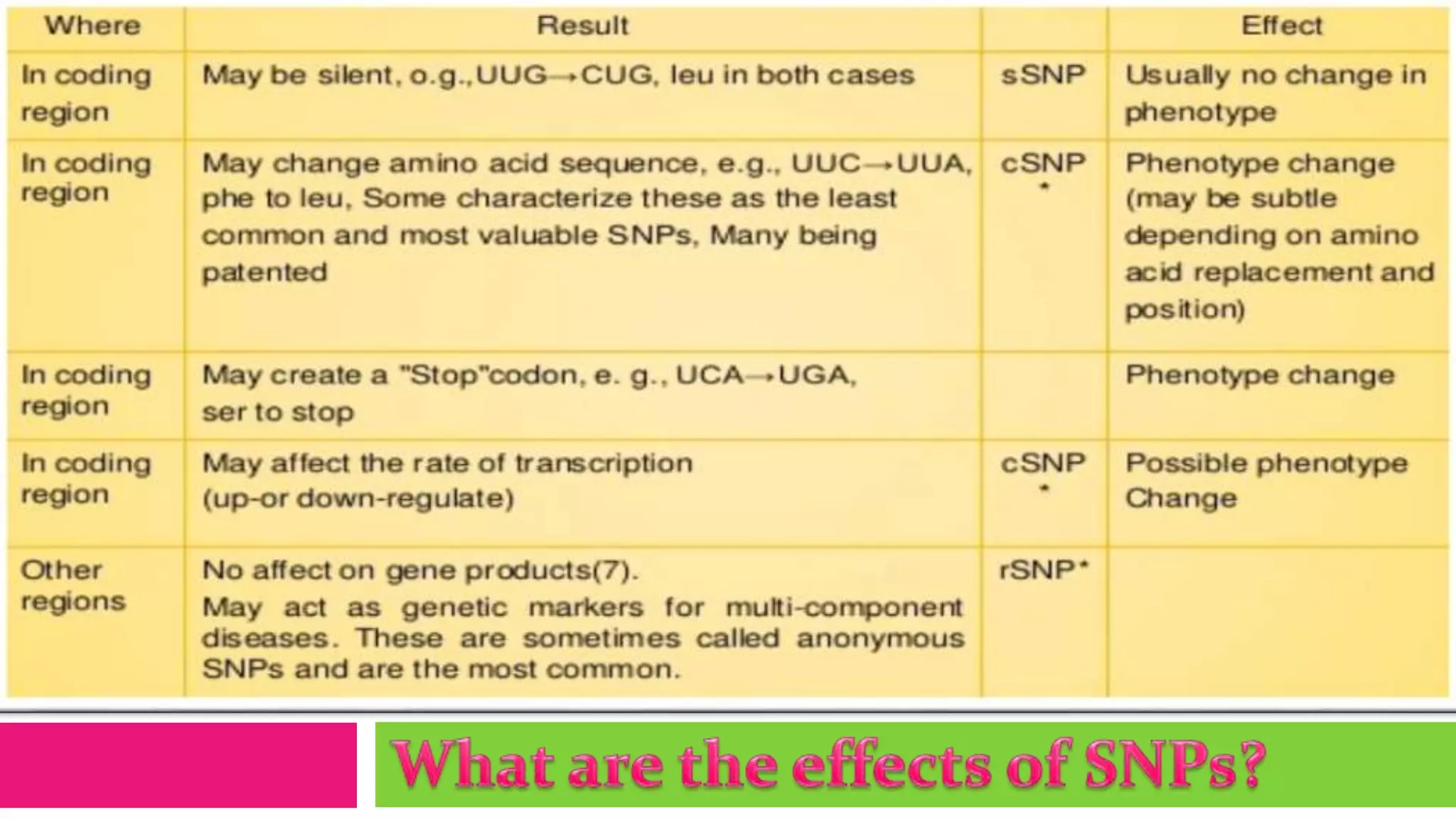
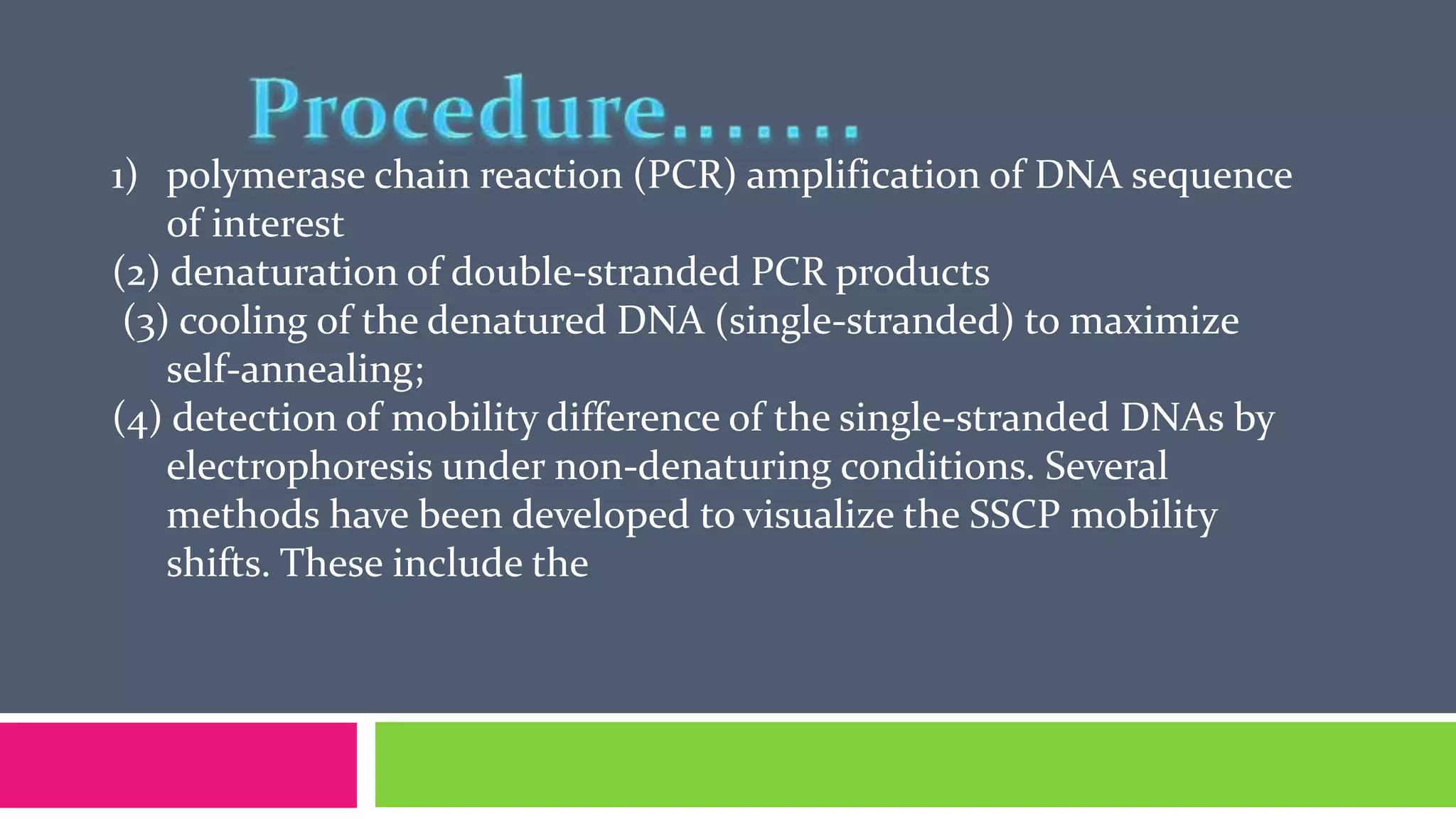
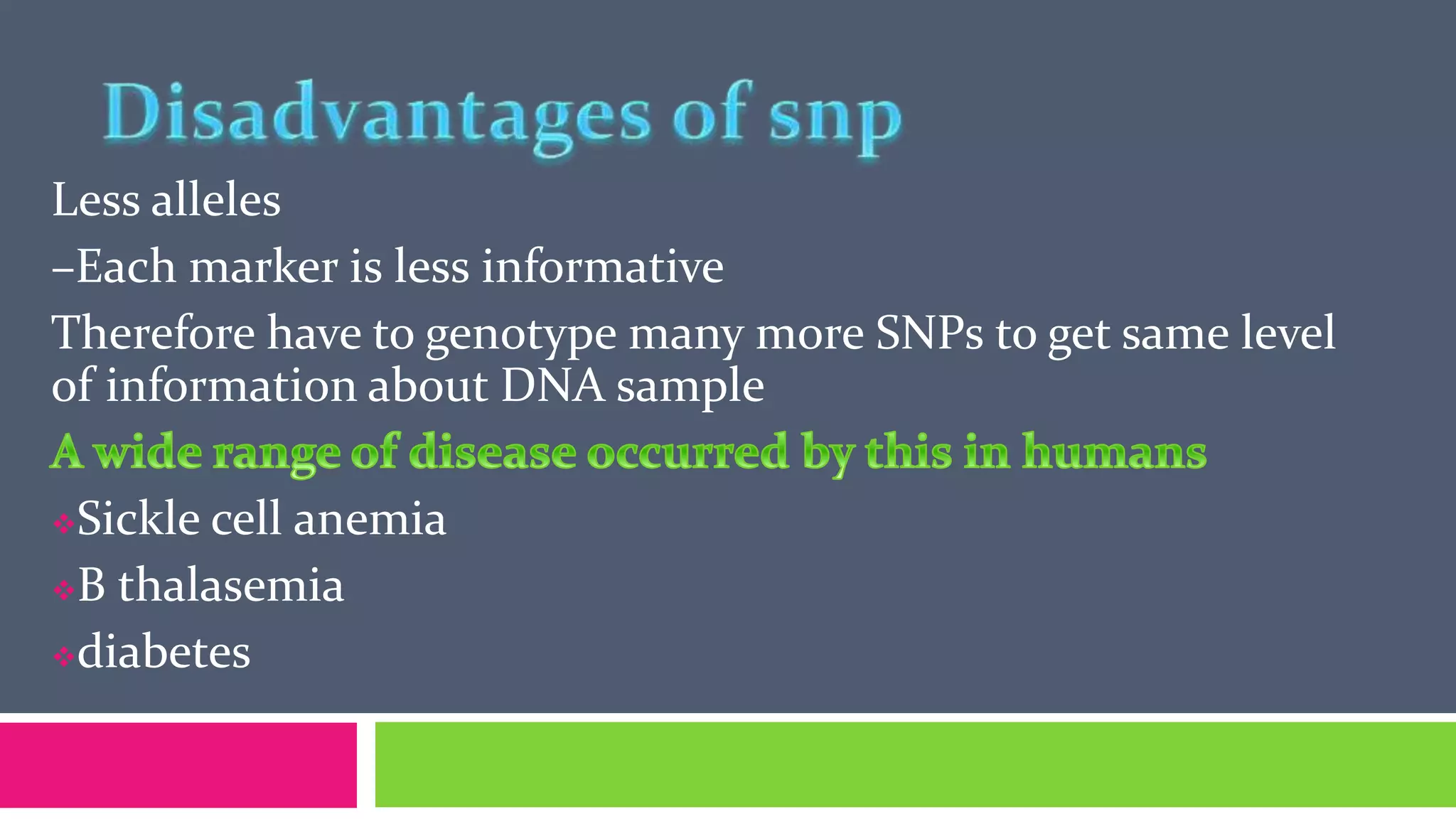
Single nucleotide polymorphisms (SNPs) are common DNA sequence variations that can influence individual traits and disease susceptibility, accounting for 0.1% of genetic diversity among humans. Research utilizing SNP profiles is crucial for understanding drug responses and developing personalized treatments, with SNPs serving as markers for genes associated with specific phenotypes. Various methods, including PCR and mass spectrometry, are employed to analyze SNPs and their impact on genetic disorders and therapeutic efficacy.