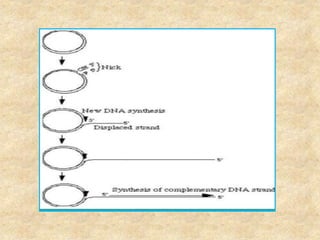
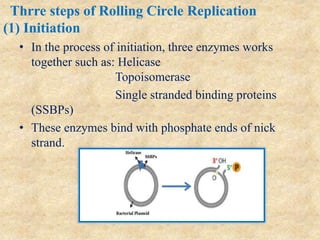
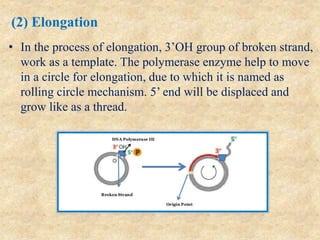
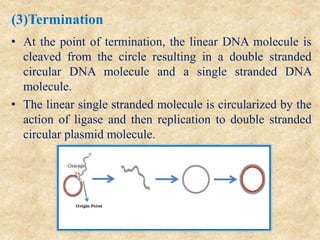
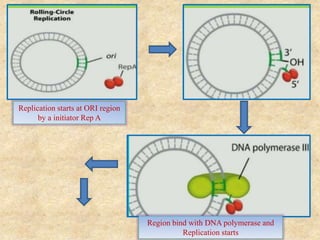
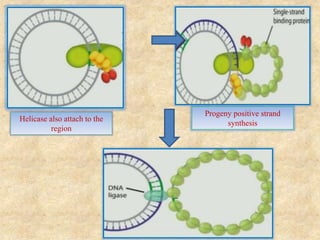
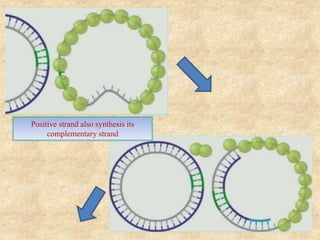
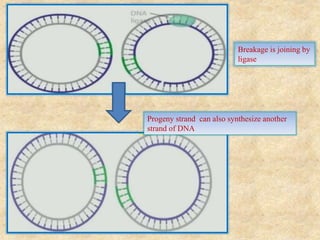
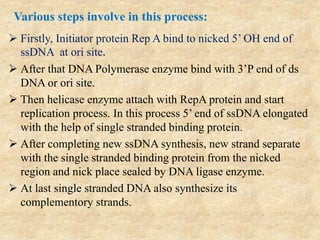
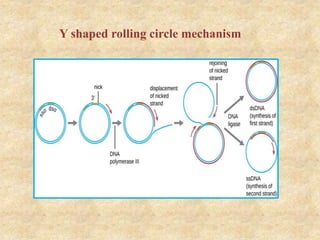
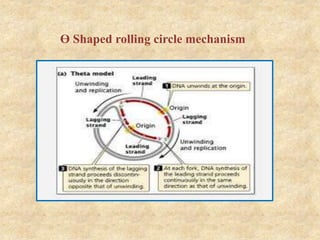
Rolling circle replication is a process that can rapidly synthesize multiple copies of circular DNA or RNA molecules. It involves the unidirectional replication of circular nucleic acids. The process begins with an initiator protein nicking one strand of the circular DNA. DNA polymerase then uses the 3' end of the nicked strand to initiate replication, displacing the 5' end. Replication continues around the circle to produce a long concatemer of copies. The concatemer is then cleaved and ligated to form multiple double-stranded circular DNA molecules. Rolling circle replication is used by some viruses and plasmids to replicate their genomes and can be harnessed for applications like signal amplification in biosensing.