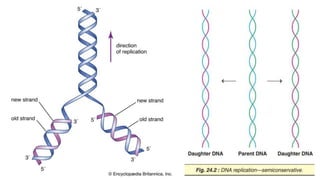
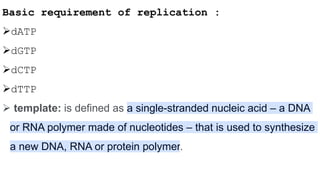
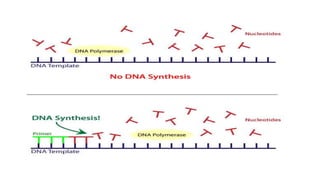
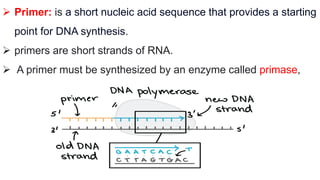
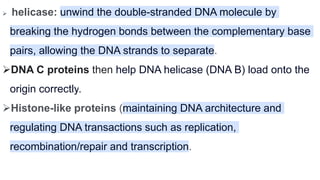
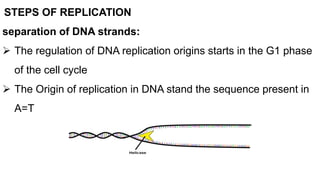
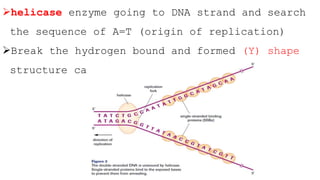
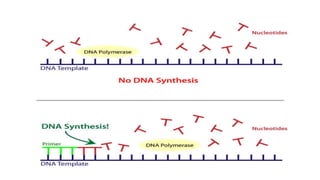
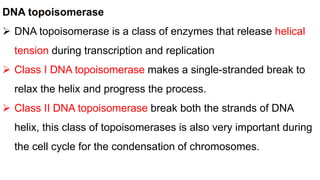
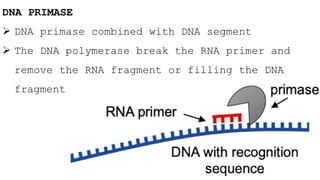
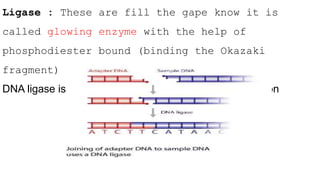
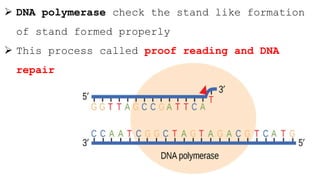
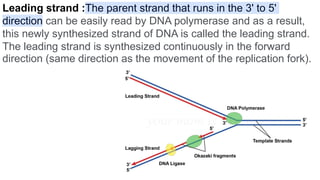
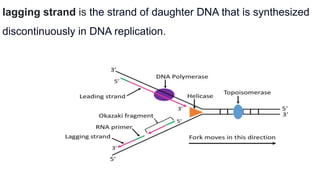
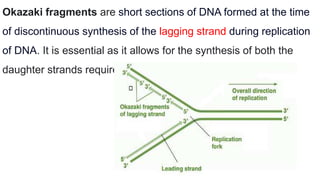
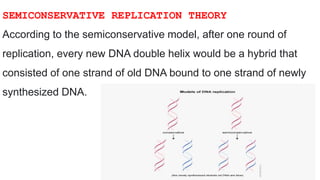
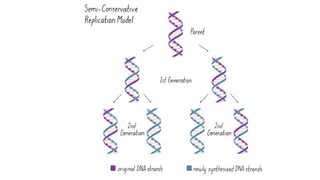
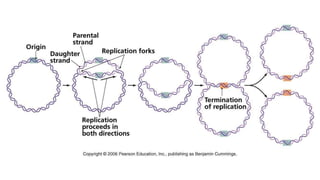
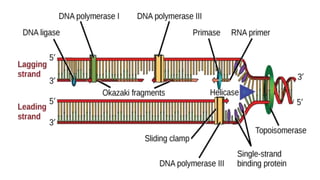
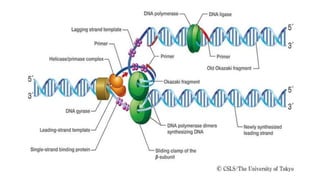
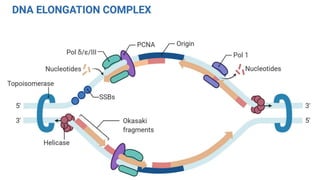
DNA replication is a critical biological process that produces identical DNA replicas for inheritance in all living organisms. It involves key components such as helicase for unwinding DNA, DNA polymerase for nucleotide addition, and ligase for sealing gaps, along with distinct processes for prokaryotes and eukaryotes. The process is semiconservative, ensuring that each new DNA molecule consists of one old strand and one newly synthesized strand.