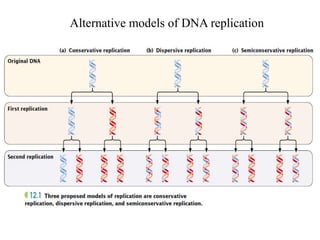
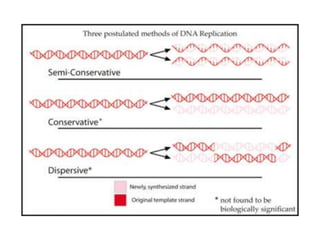
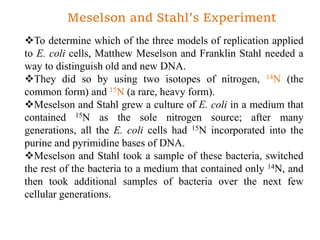
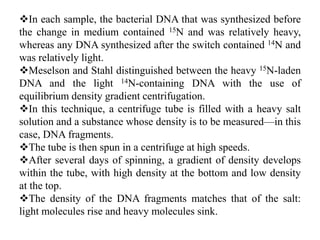
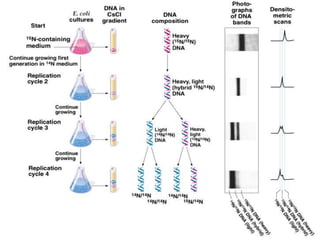
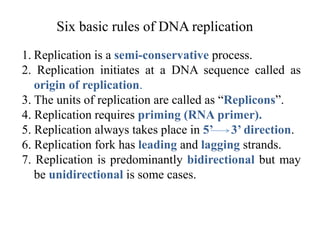
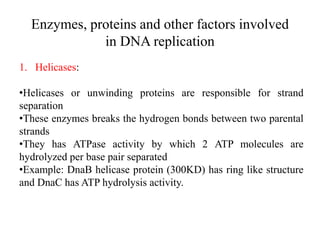
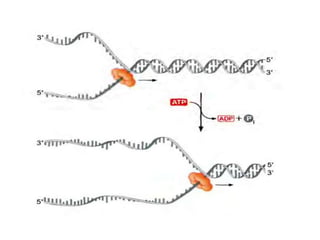
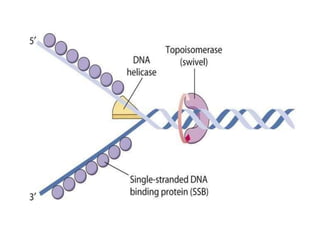
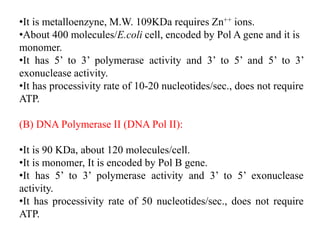
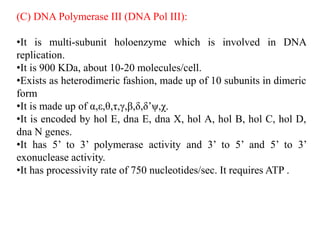
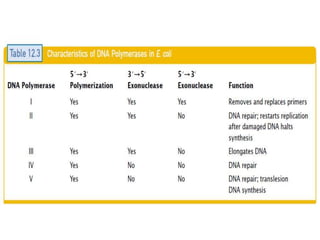
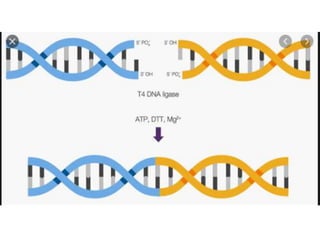
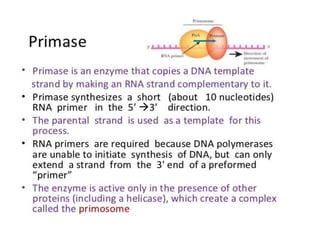
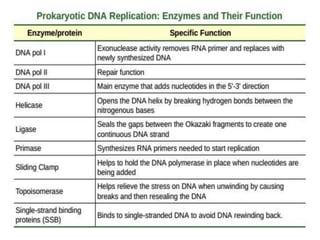
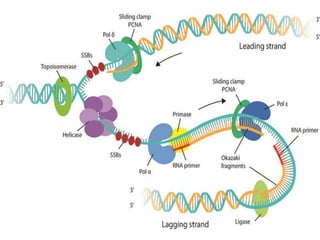
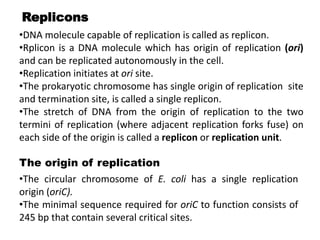
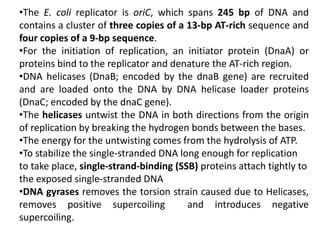
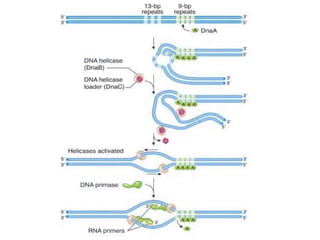
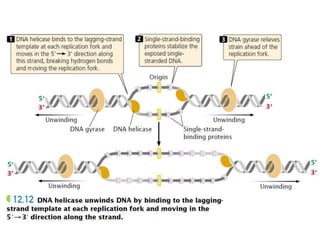
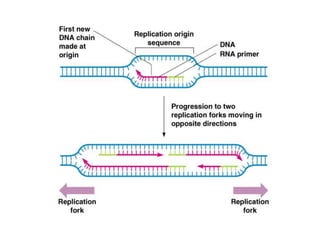
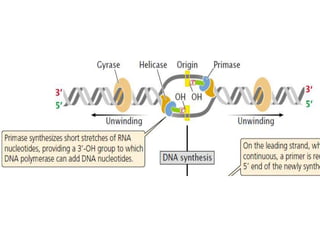
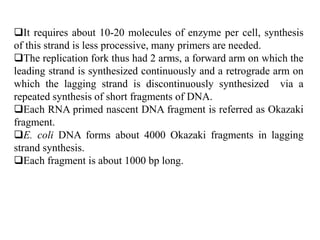
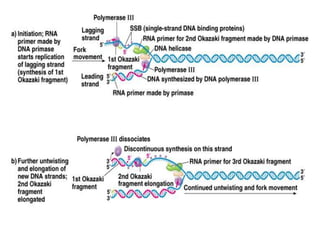
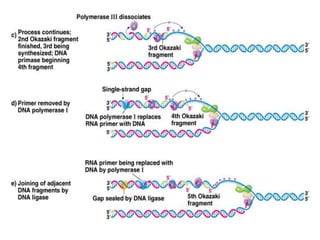
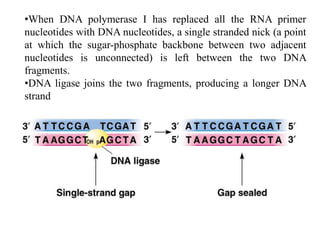
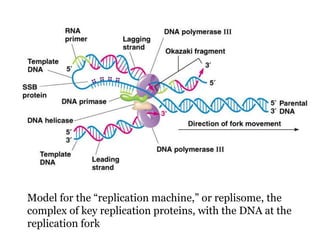
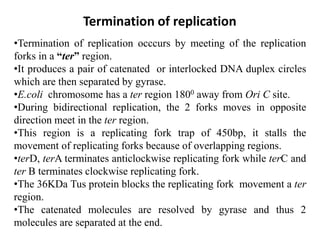
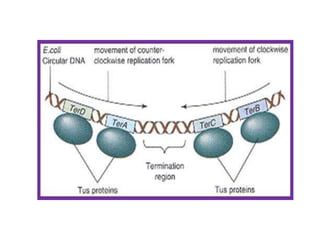
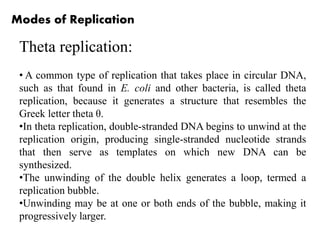
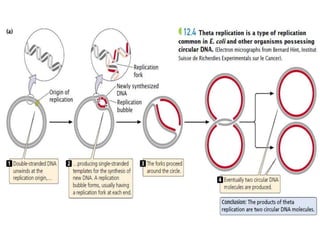
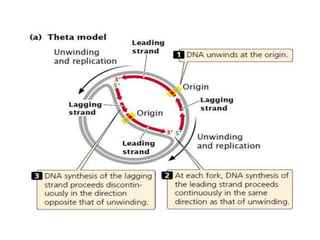
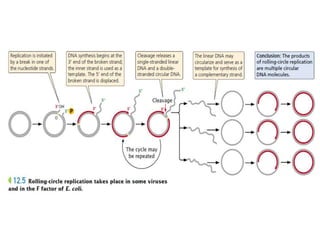
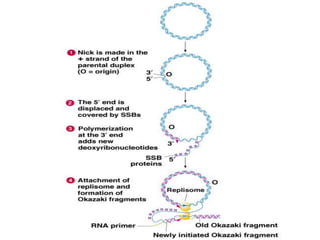
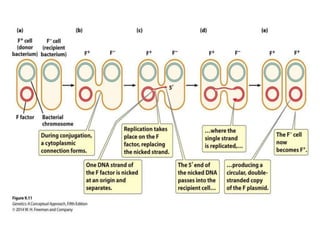
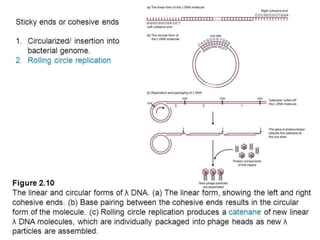
The document discusses prokaryotic DNA replication, highlighting different models such as conservative, semiconservative, and dispersive replication, with a focus on the semiconservative model proven by Meselson and Stahl's experiment. It outlines key components, including the enzymes involved (helicases, topoisomerases, DNA polymerases, and ligases), and describes the process of replication initiation, strand synthesis, and the roles of RNA primers and Okazaki fragments in lagging strand synthesis. Additionally, it emphasizes the significance of the origin of replication and provides a detailed look at the mechanics of DNA polymerization and the leading and lagging strand dynamics.