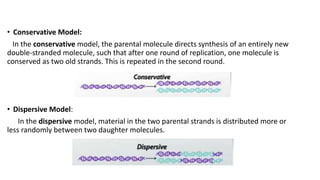
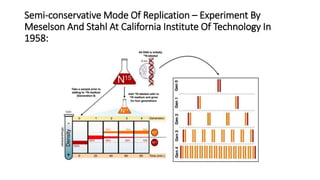
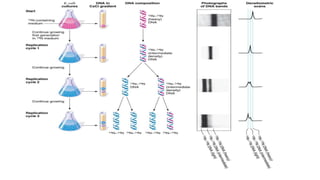
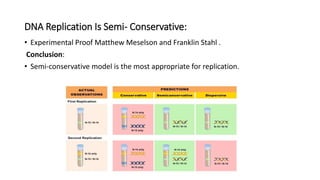
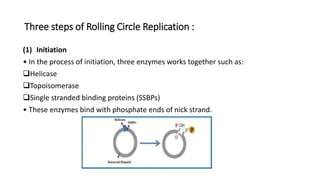
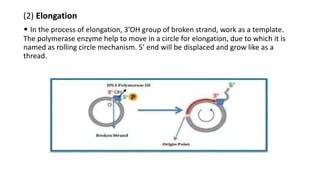
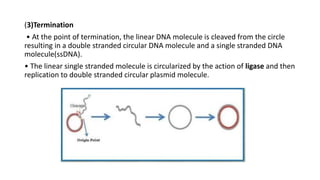
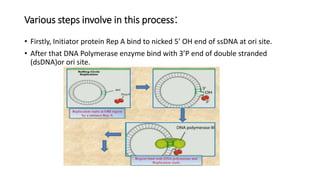
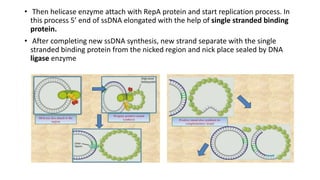
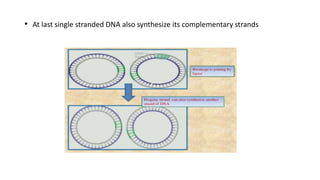
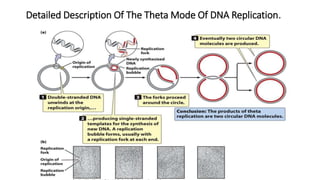
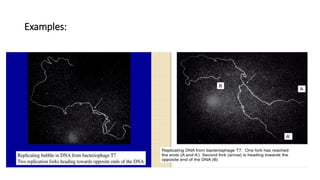
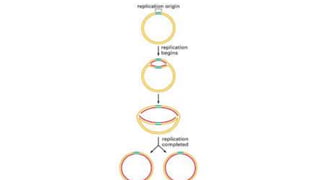
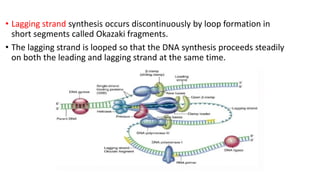
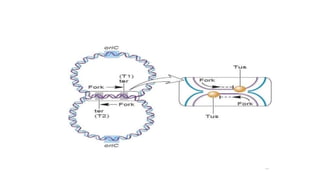
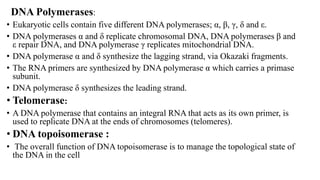
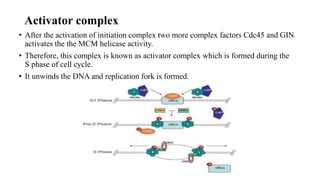
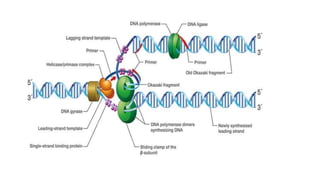
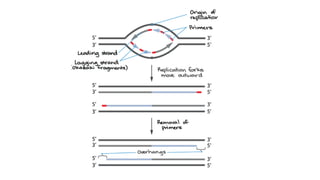
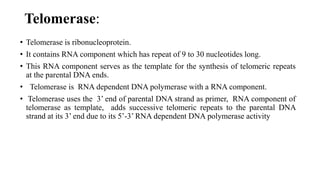
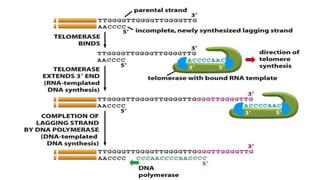
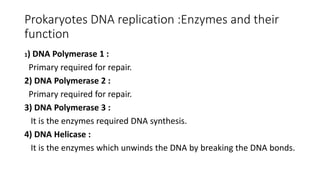
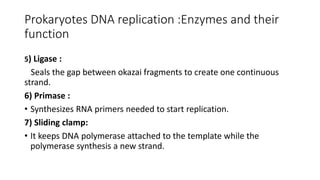
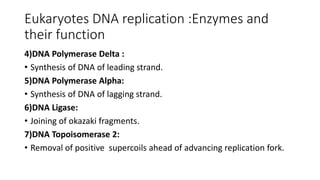
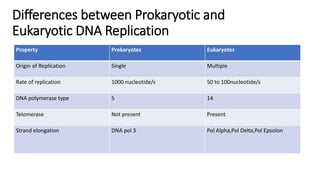
This document discusses DNA replication in prokaryotes and eukaryotes. It provides an overview of the key steps and enzymes involved in DNA replication for both prokaryotes and eukaryotes. For prokaryotes, it describes initiation at the origin of replication involving DNA A protein, elongation by DNA polymerase III, and termination when replication forks meet. For eukaryotes, it outlines initiation involving pre-replication complexes, elongation involving leading and lagging strand synthesis, and the various enzymes involved such as DNA polymerases and helicases.