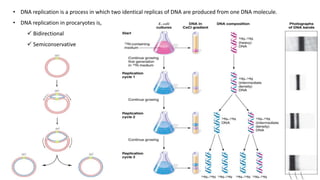
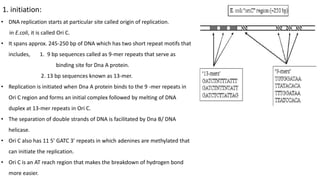
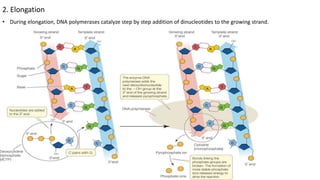
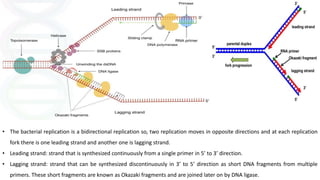
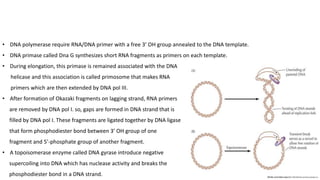
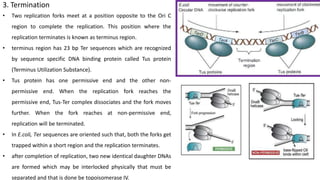
DNA replication in prokaryotes occurs through three main steps: initiation, elongation, and termination. Initiation begins at the origin of replication when DnaA and other proteins help separate the DNA strands. Elongation then uses DNA polymerases and other enzymes to bidirectionally copy the DNA in a semiconservative manner, producing both a leading and lagging strand. Termination occurs when the replication forks meet at the terminus region, utilizing proteins like Tus to stop replication.