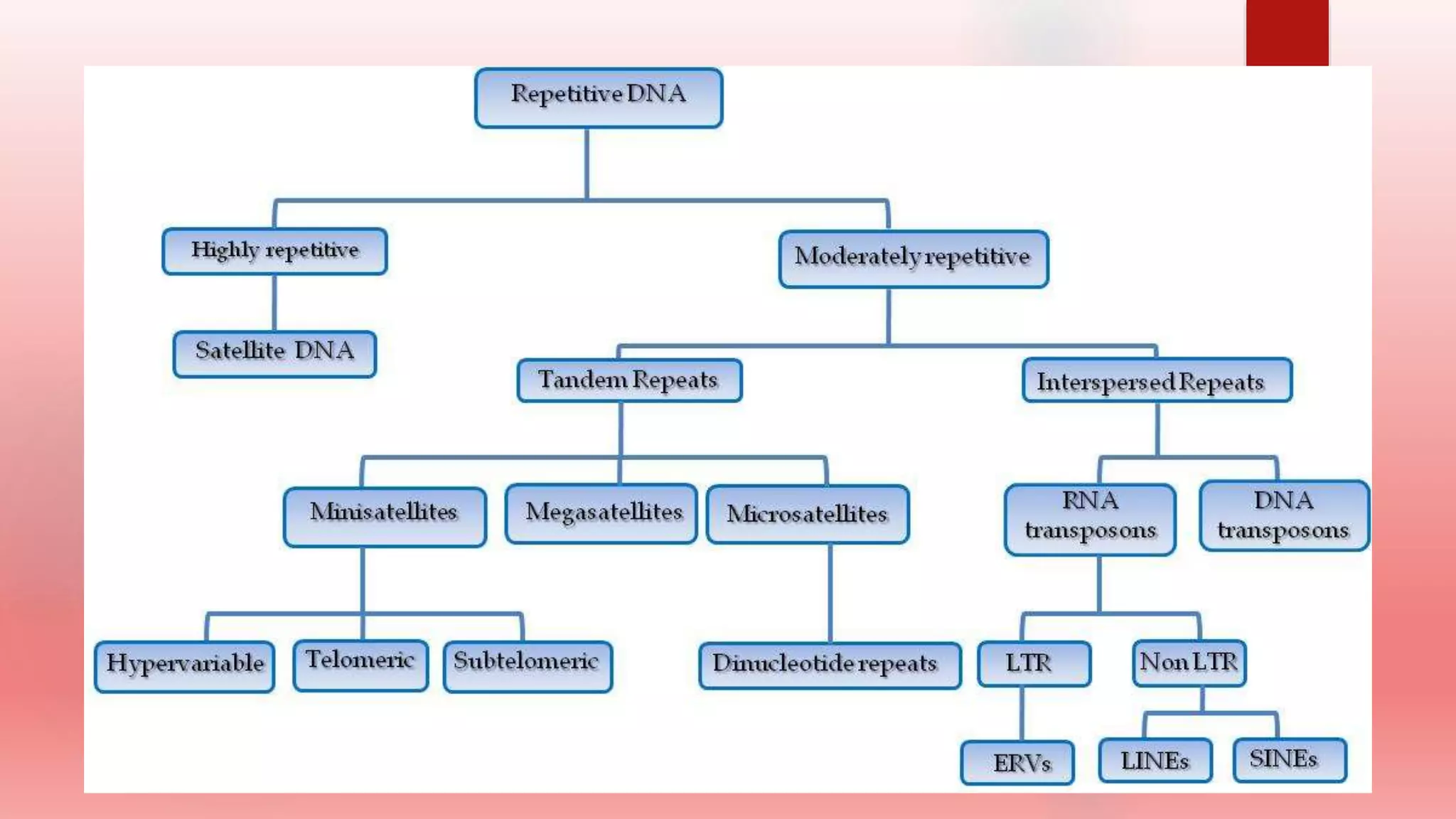
The document discusses DNA methylation, repetitive DNA sequences, the C-value paradox, and their relationships. It provides background on DNA methylation, how it regulates gene expression and is involved in diseases. It describes highly repetitive and satellite DNA sequences. It explains that the C-value paradox stemmed from the observation that genome size did not correlate with complexity, but this was later resolved by discovering non-coding DNA. The paradox questioned how genome size related to gene number.