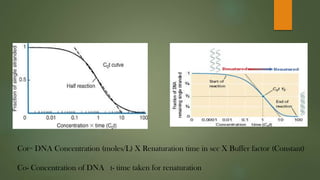
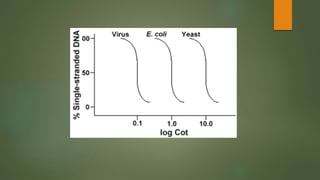
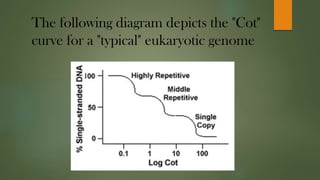
The document describes Cot curve analysis, a technique used to determine DNA sequence complexity in genomes. DNA is denatured by heating and allowed to renature at different time points. The percentage of single-stranded DNA is plotted against Cot value, the product of DNA concentration and renaturation time. Species with more complex genomes have higher Cot1/2 values as complementary sequences take longer to encounter each other. Eukaryotic genomes contain repetitive sequences that affect the Cot curve shape, with highly repetitive sequences renaturing fastest and single-copy sequences slowest.