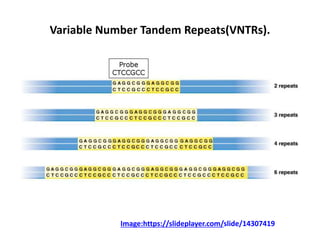
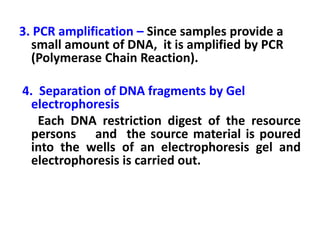
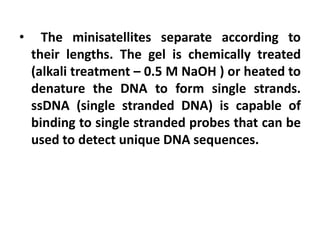
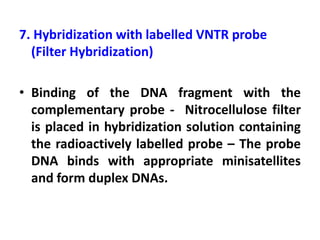
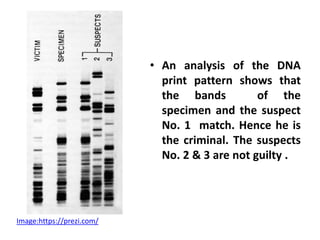
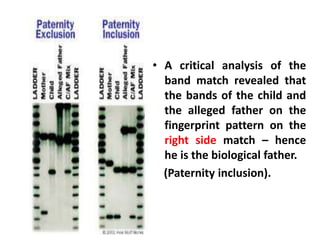
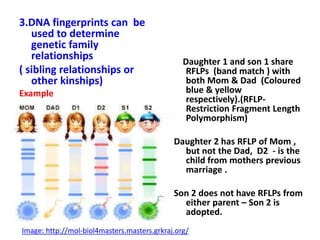
DNA fingerprinting is a technique used to identify individuals by analyzing genetic variations in DNA sequences found in certain regions of the genome. It was developed in 1985 by Alec Jeffreys and involves isolating DNA from a sample, cutting it with restriction enzymes, and comparing patterns of DNA fragments between individuals using probes that bind to variable number tandem repeats. DNA fingerprinting relies on differences in repetitive sequences that vary between people but are identical in each cell of an individual, allowing a unique genetic profile to be generated for identification purposes in forensic investigations and paternity testing.