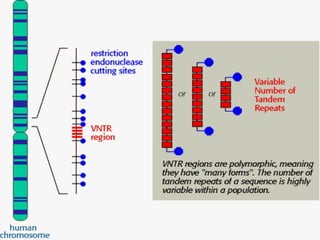
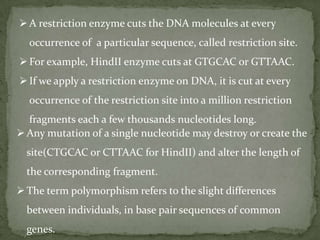
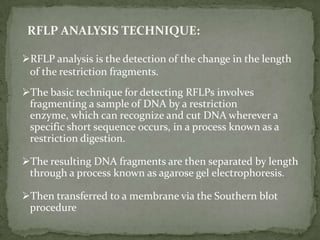
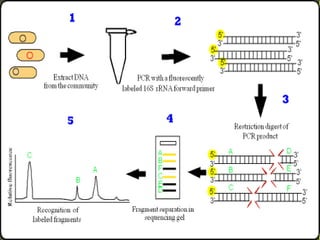
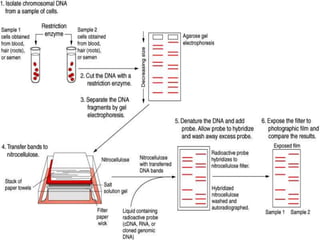
DNA fingerprinting is a method used to identify individuals based on variations in their DNA sequences. It involves using restriction enzymes to cut DNA into fragments at specific sequences, which are then separated by size using gel electrophoresis. The pattern of fragment sizes is unique to each individual and can be used to identify a person, determine paternity, solve criminal cases, diagnose genetic disorders, and develop cures. The document provides details on the different types of tandem repeats like satellites, minisatellites, and microsatellites that contribute to variations between individuals, as well as the basic process and applications of DNA fingerprinting.