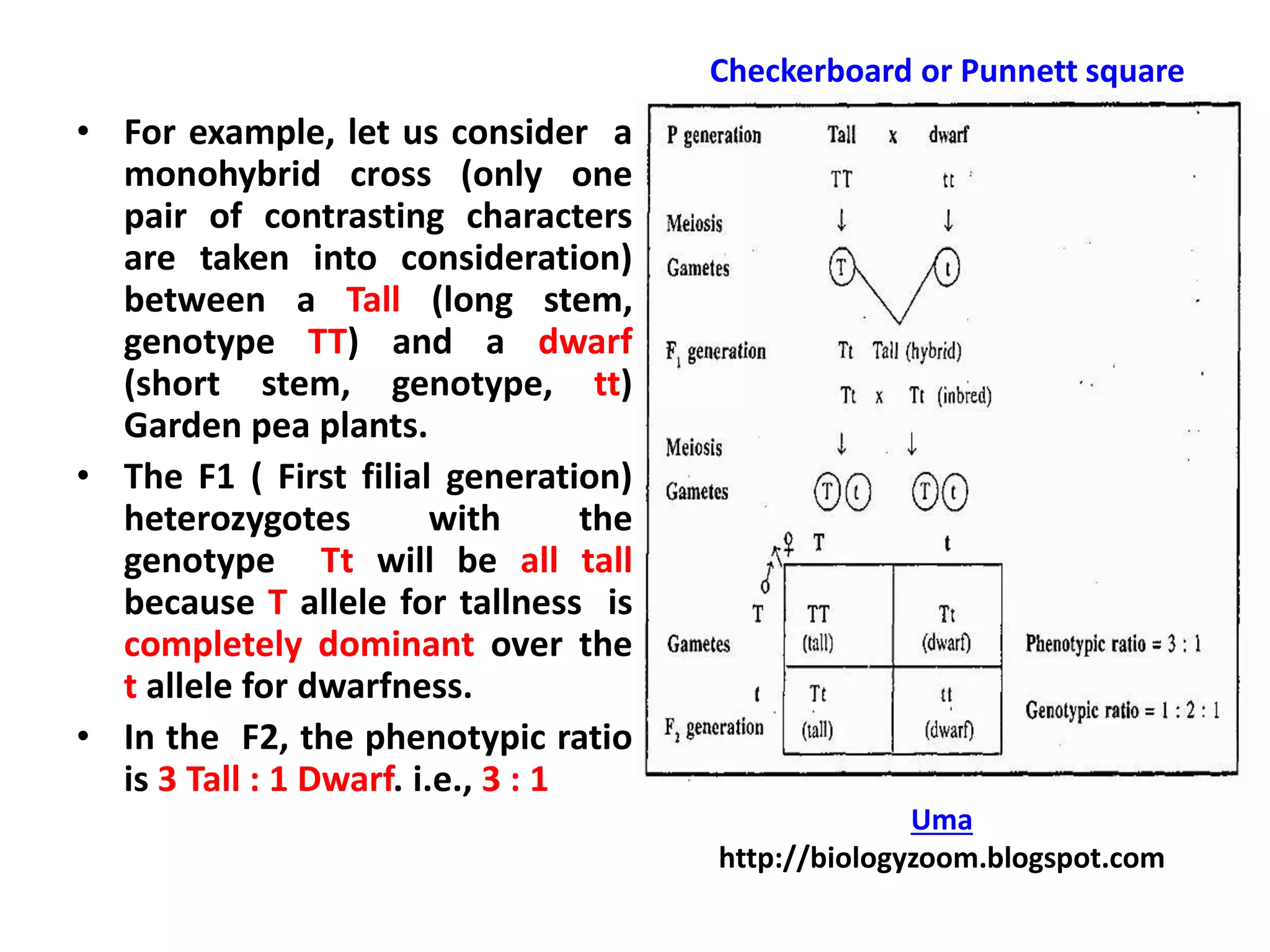
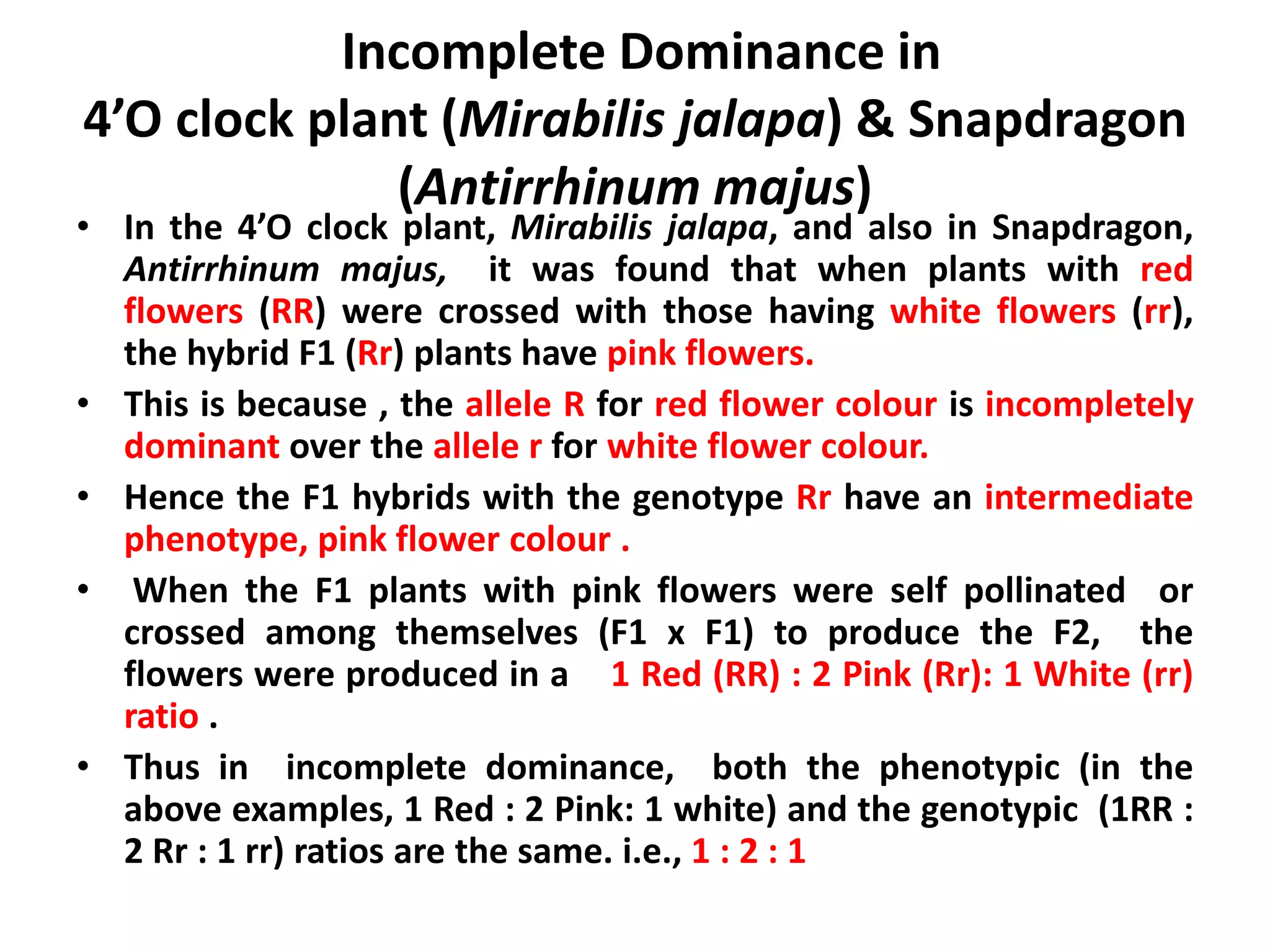
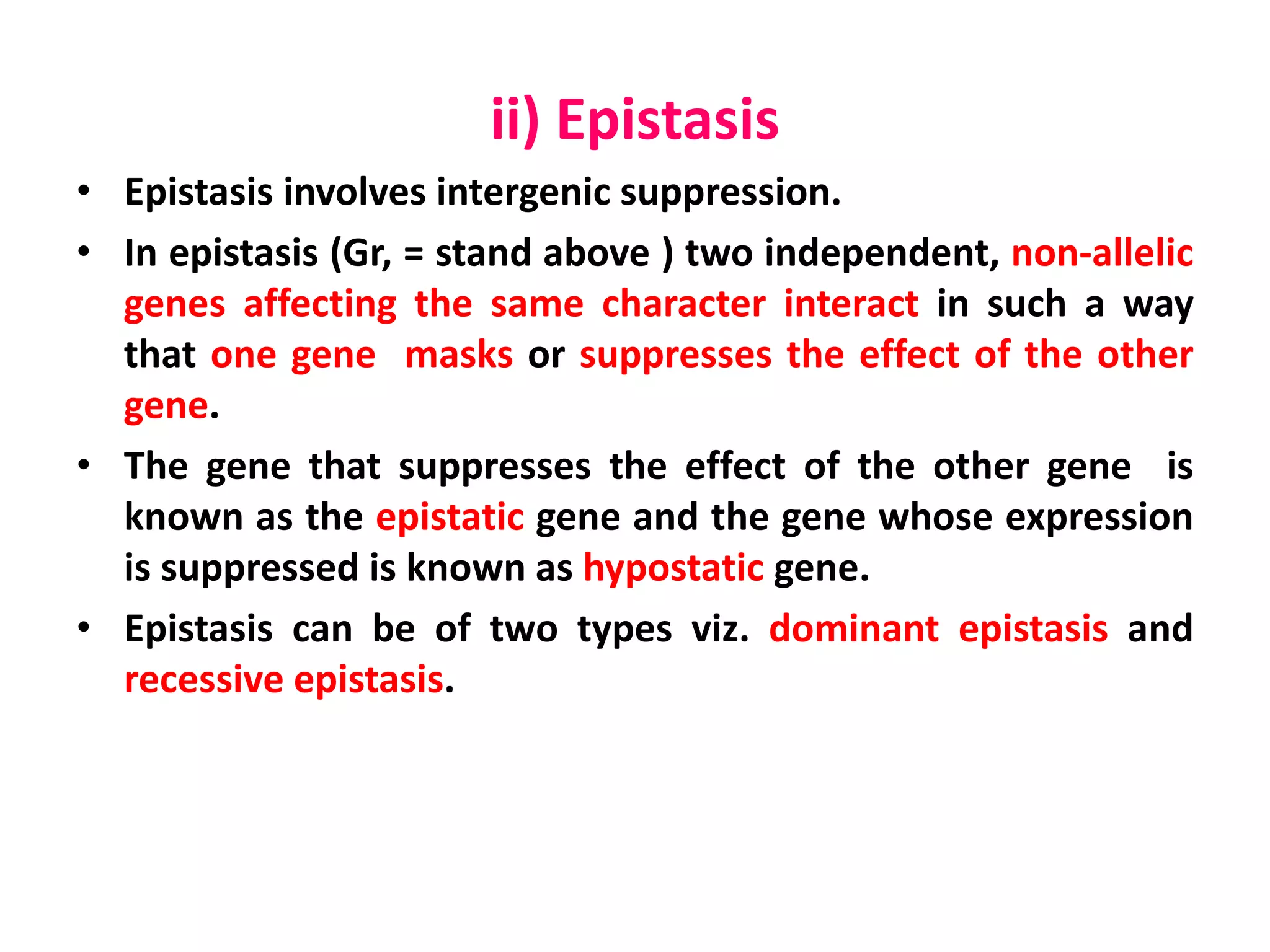
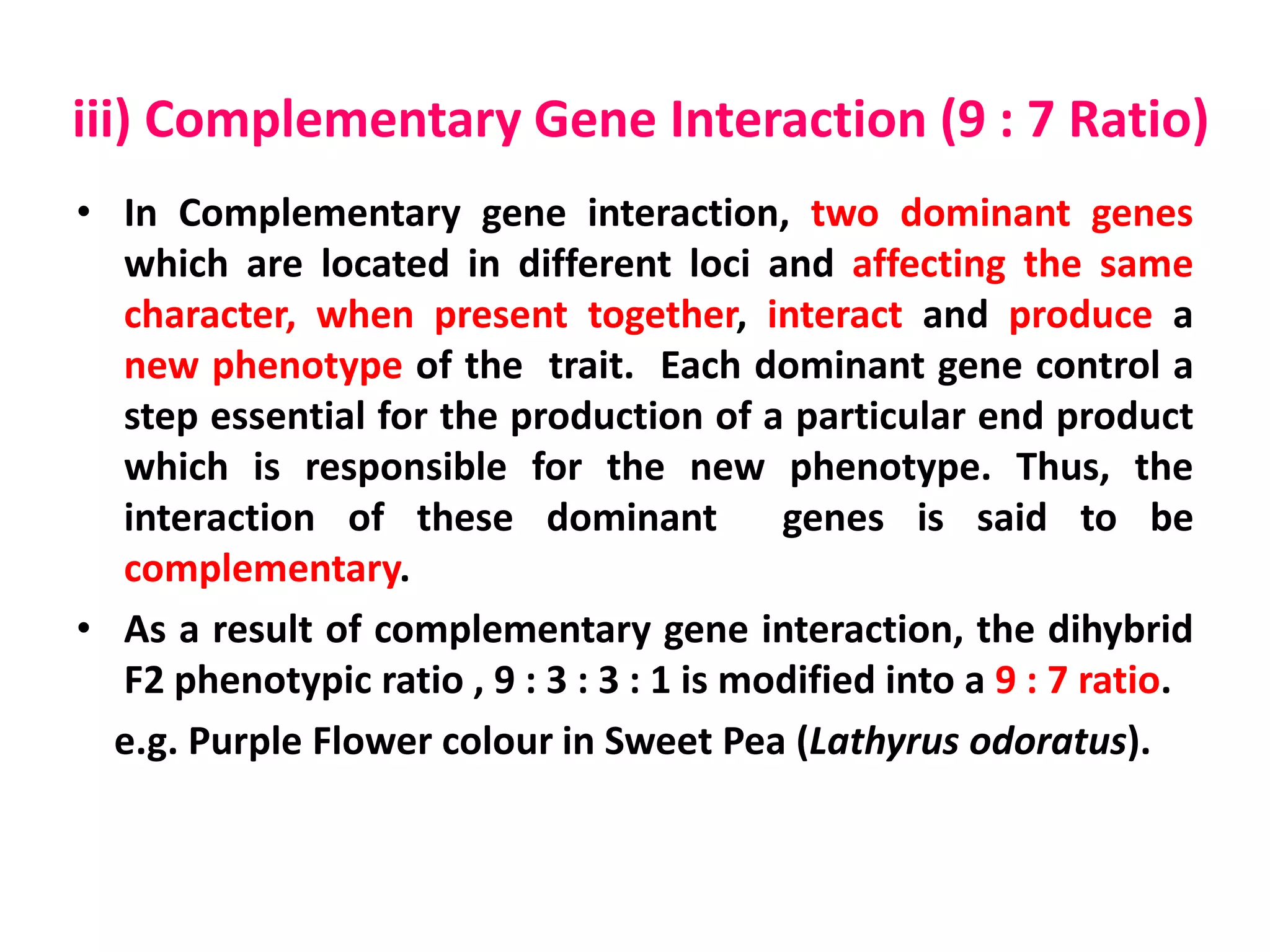
Gene interactions play an important role in inheritance and can produce phenotypes that do not follow typical Mendelian ratios. There are two main types of gene interactions - allelic and non-allelic. Allelic interactions occur between alleles of the same gene and can be complete dominance, incomplete dominance, or co-dominance. Non-allelic interactions occur between genes and can be simple interactions like those controlling comb patterns in chickens, or more complex interactions like epistasis where one gene suppresses the expression of another. Understanding gene interactions is crucial for explaining exceptions to Mendelian inheritance.