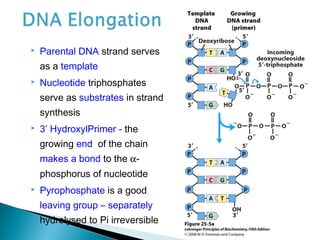
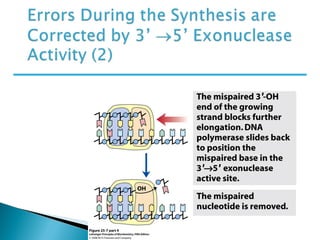
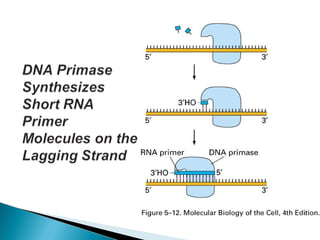
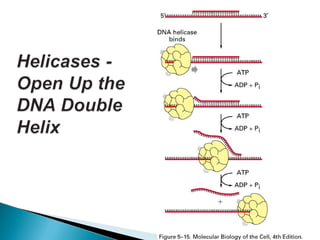
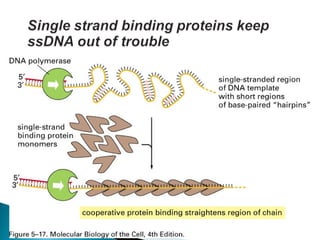
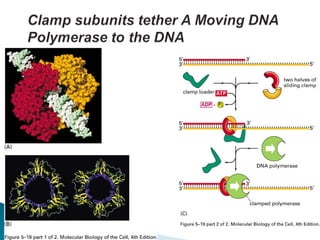
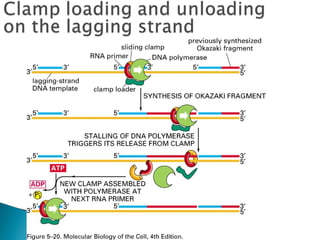
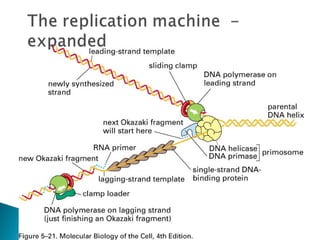
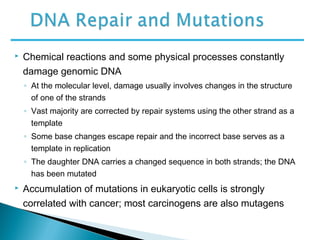
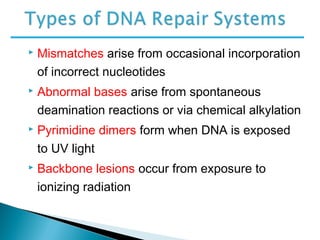
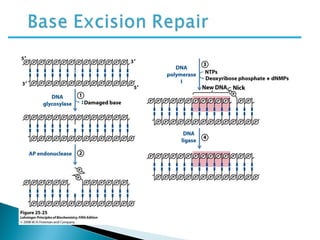
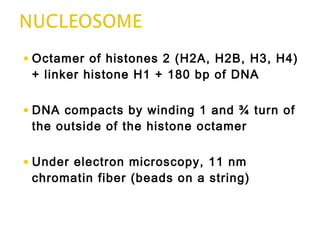
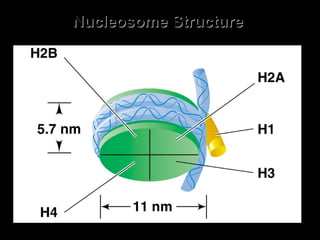
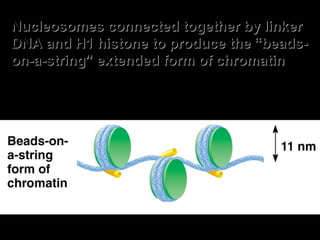
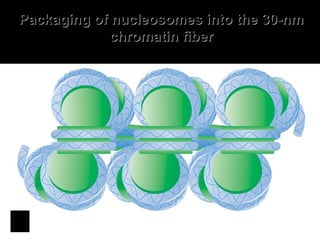
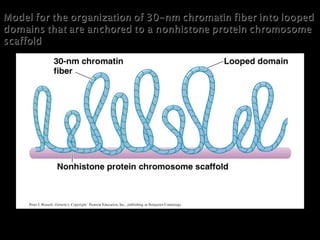
The document discusses several key topics related to DNA structure and function:
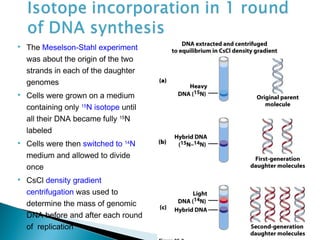
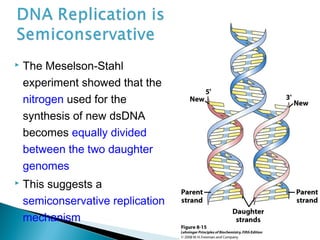
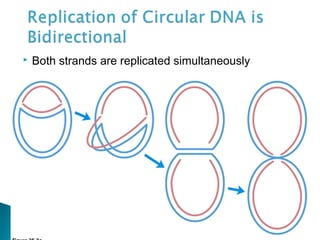
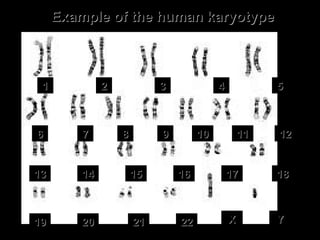
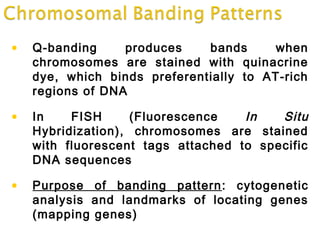
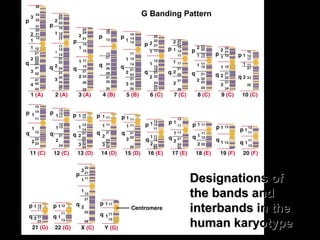
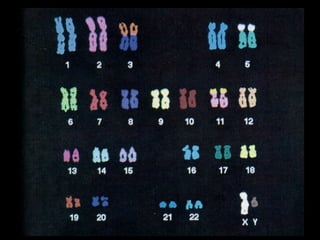
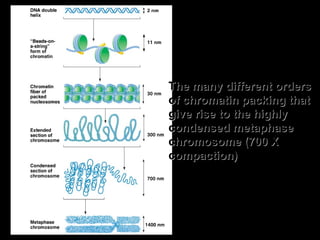
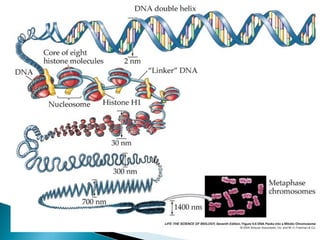
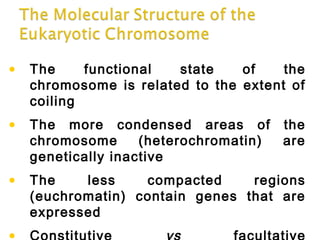
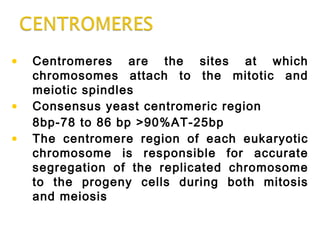
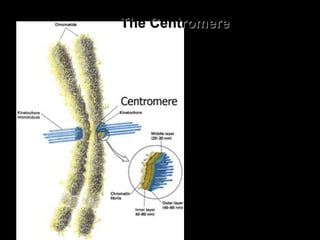
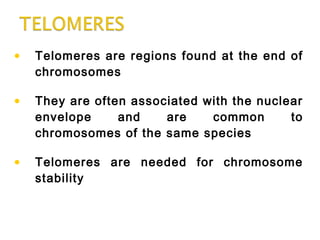
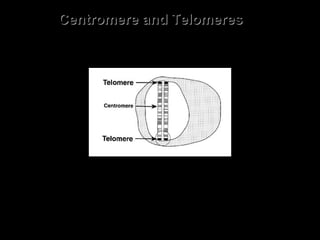
DNA replication ensures each cell has an exact copy of the DNA before cell division. Errors are constantly checked and repaired to maintain high fidelity. DNA is also rearranged through processes like recombination. The tightly regulated enzymes that perform these metabolic processes were demonstrated by the Meselson-Stahl experiment to replicate DNA using a semiconservative mechanism. DNA is organized through various levels of compaction into condensed chromosomes. This dynamic structure, along with features like centromeres and telomeres, helps regulate DNA accessibility and proper segregation during cell division.