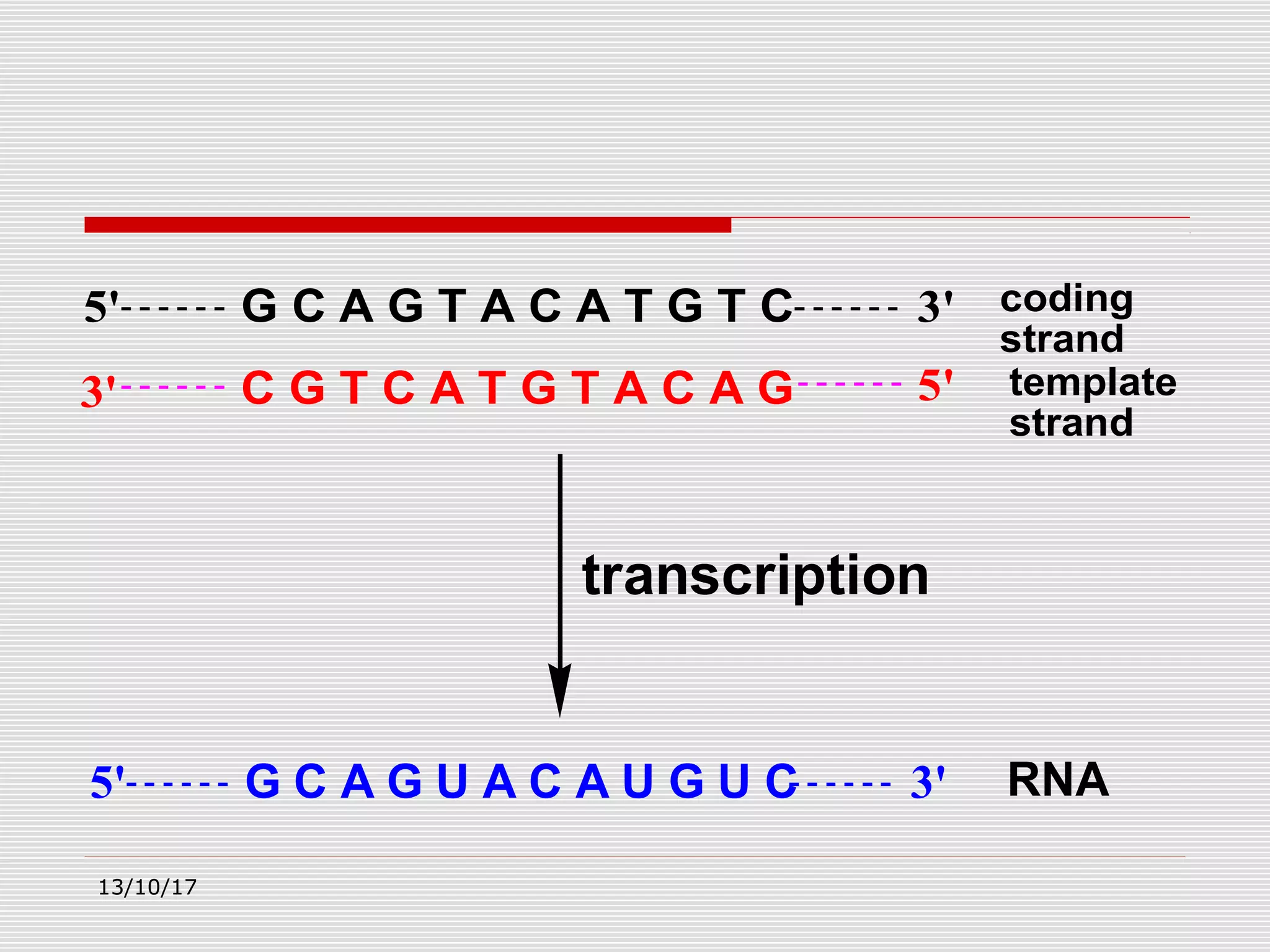
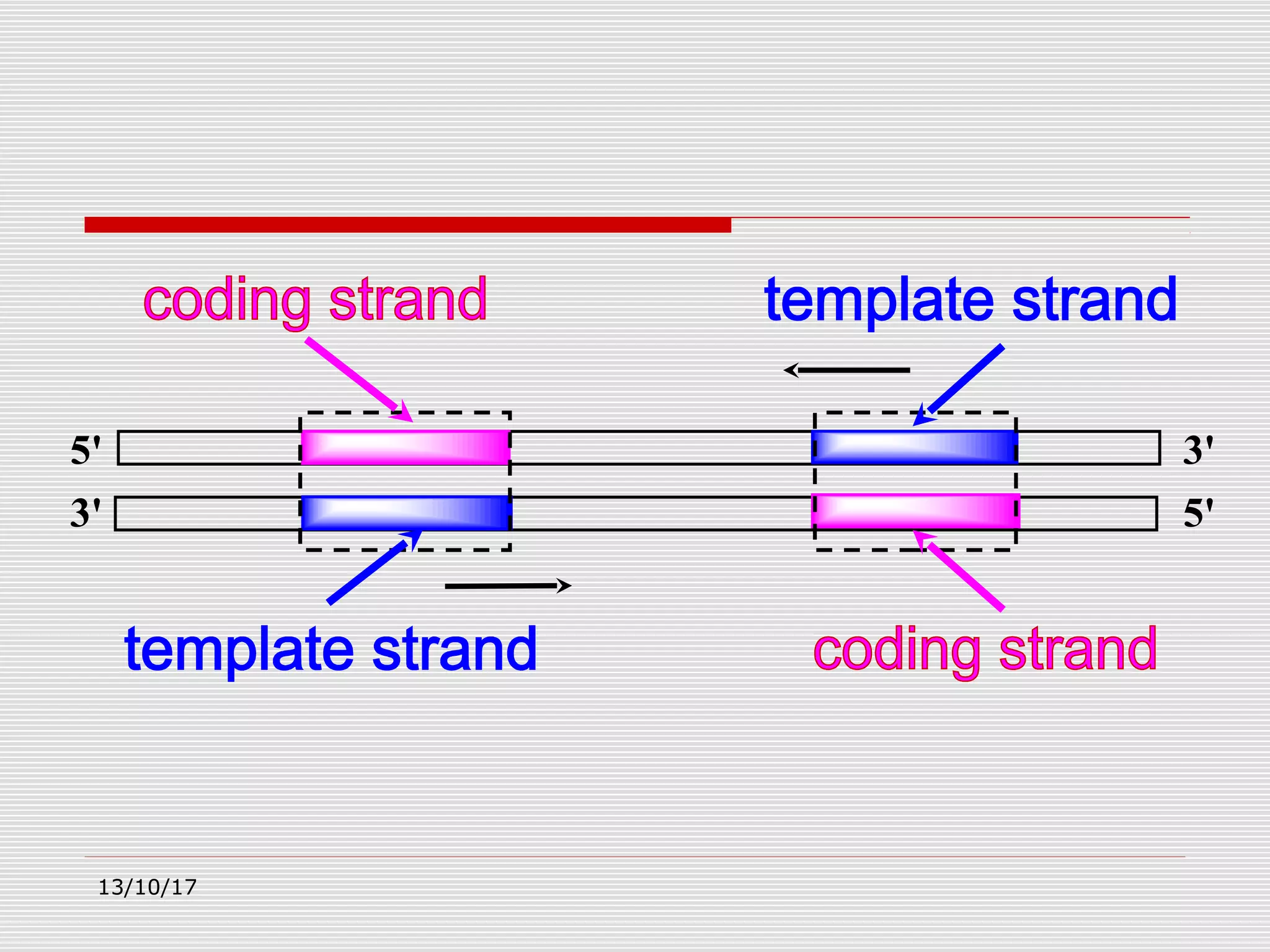
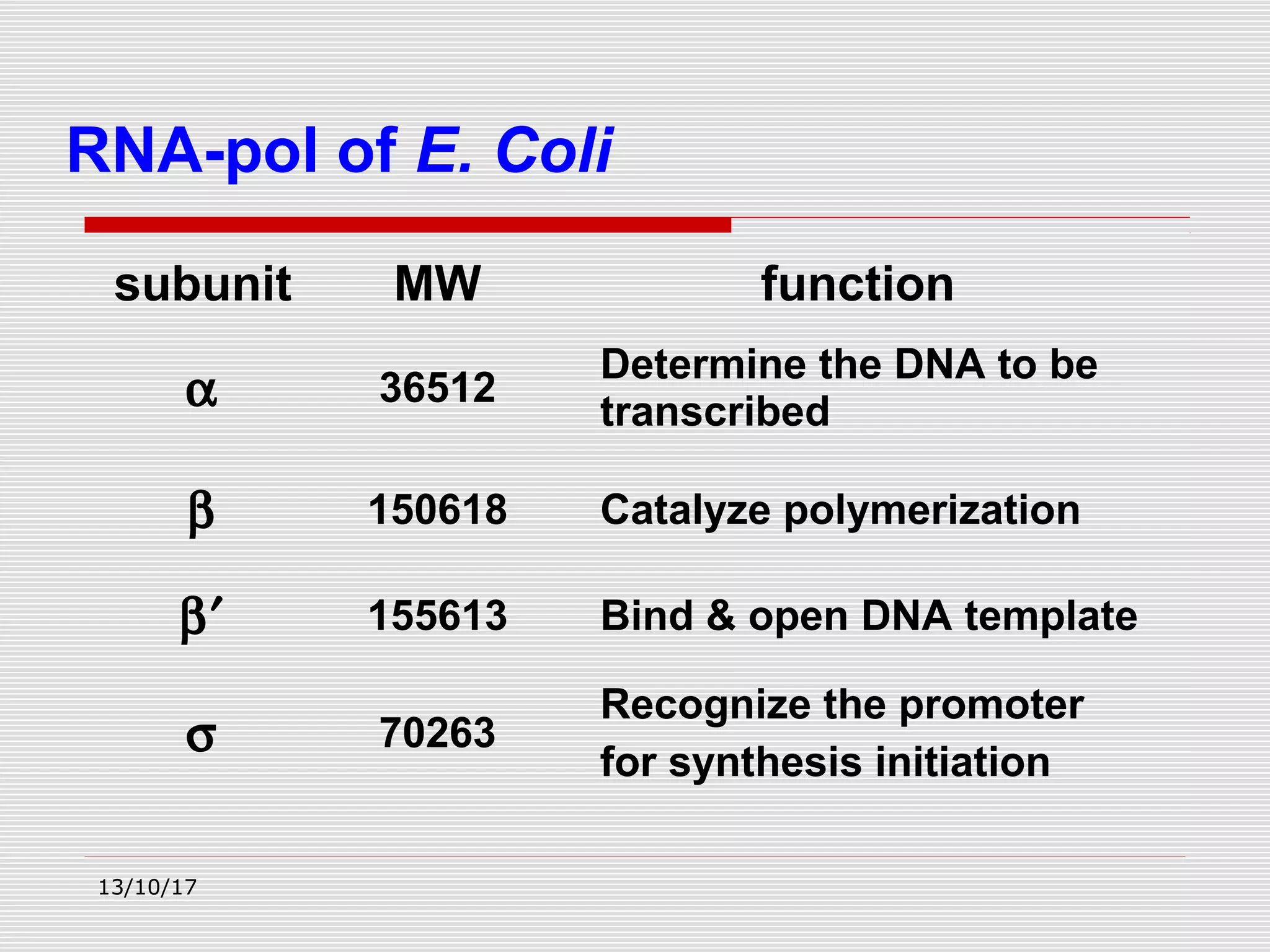
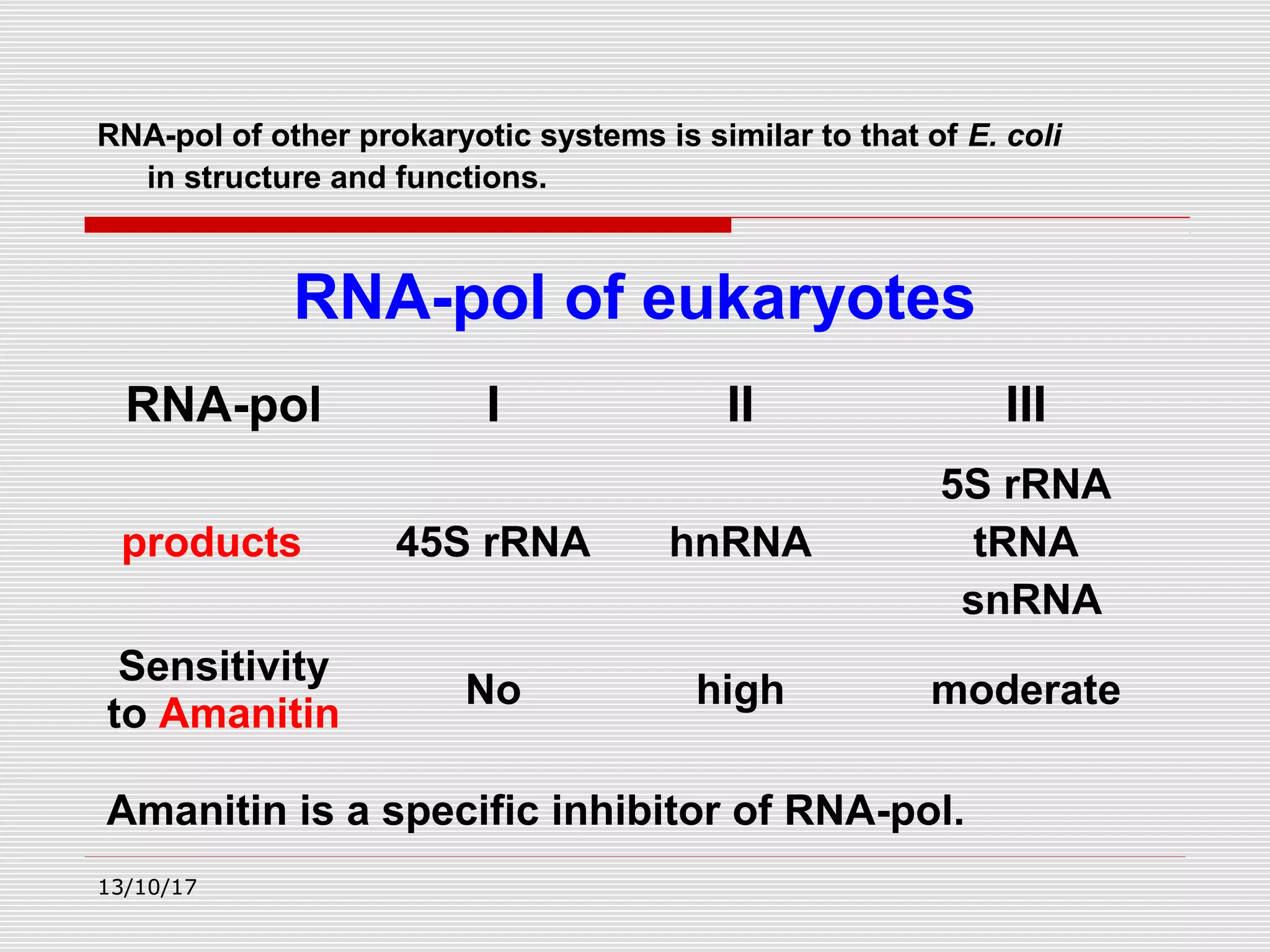
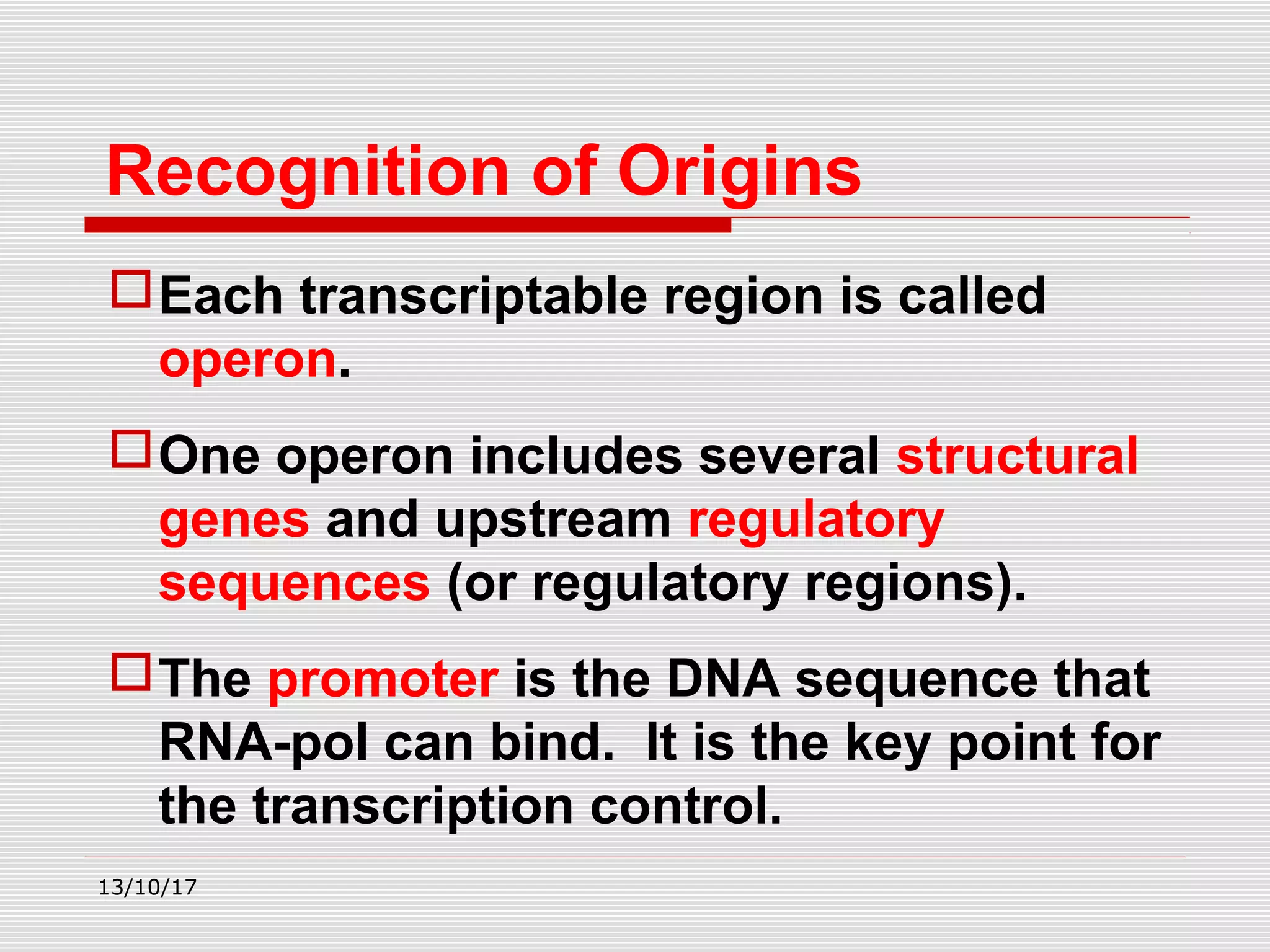
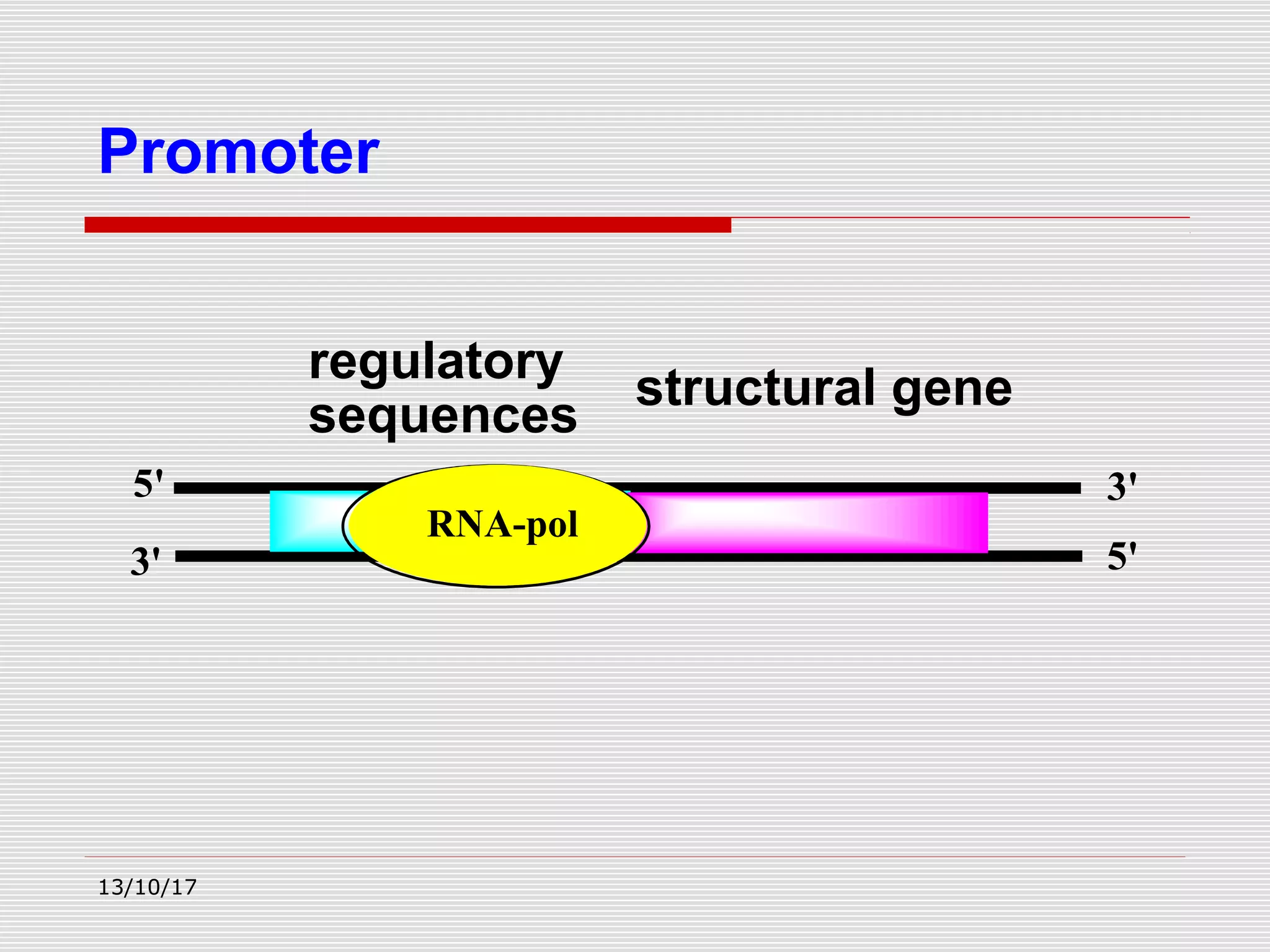
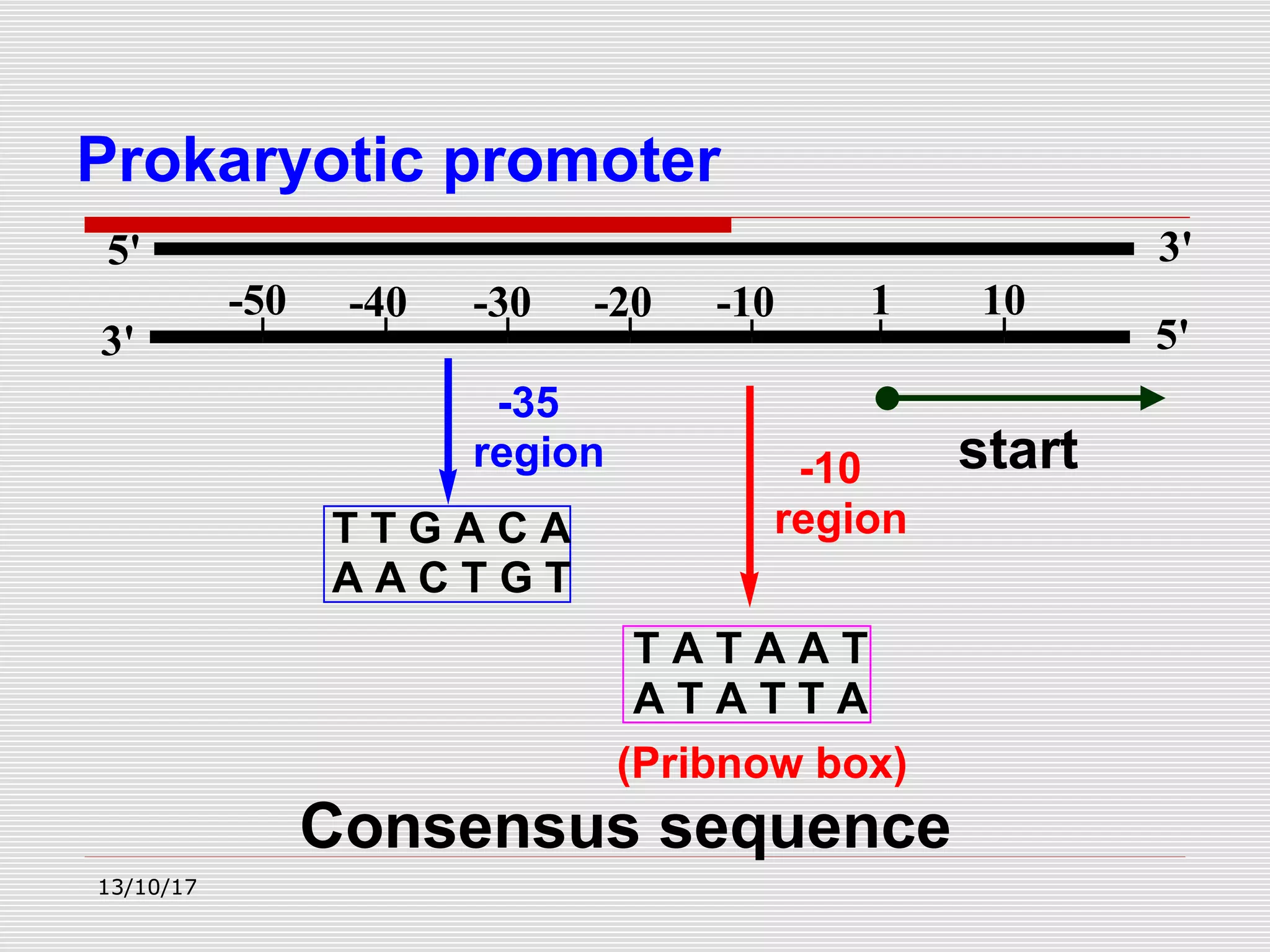
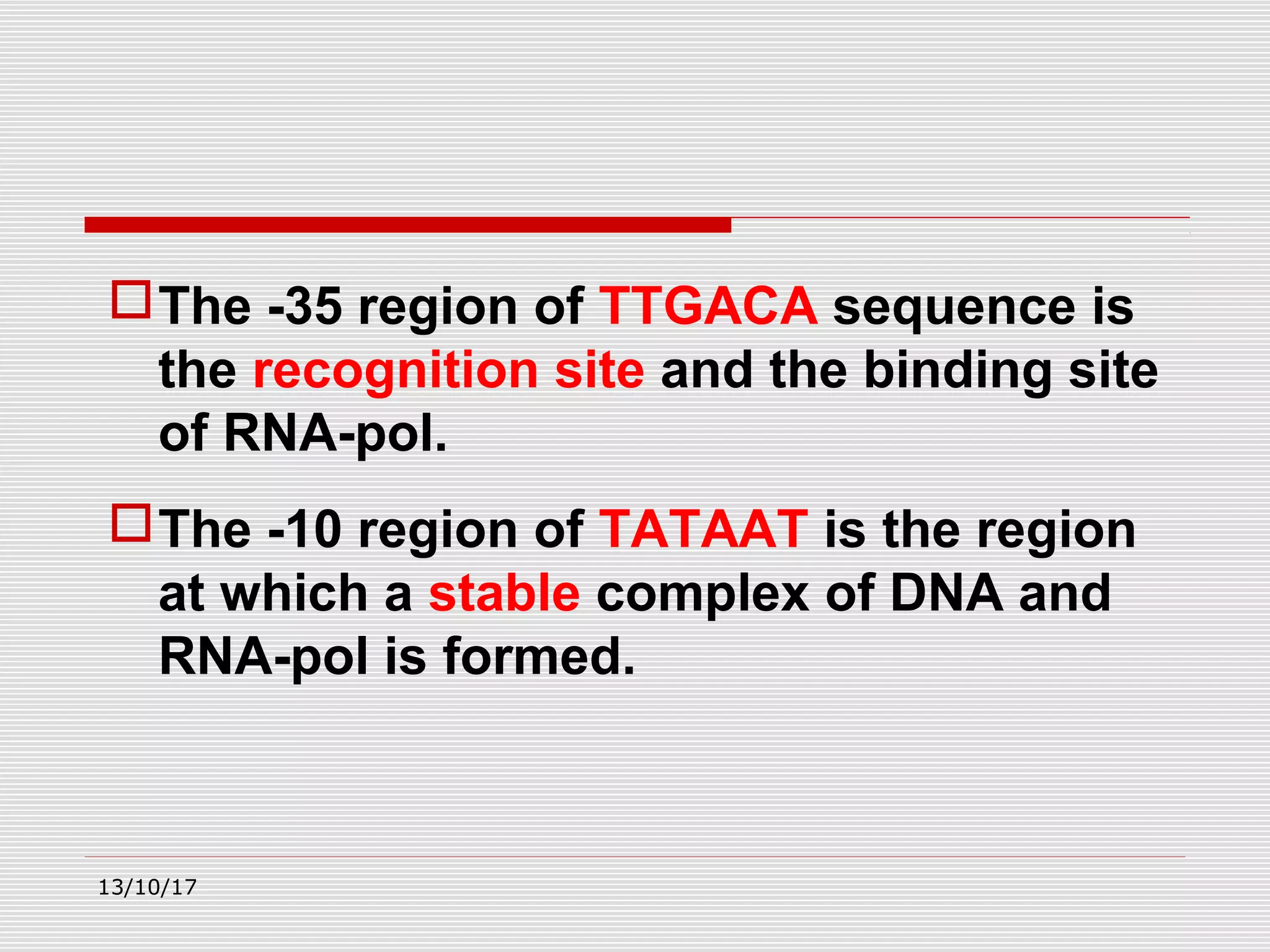
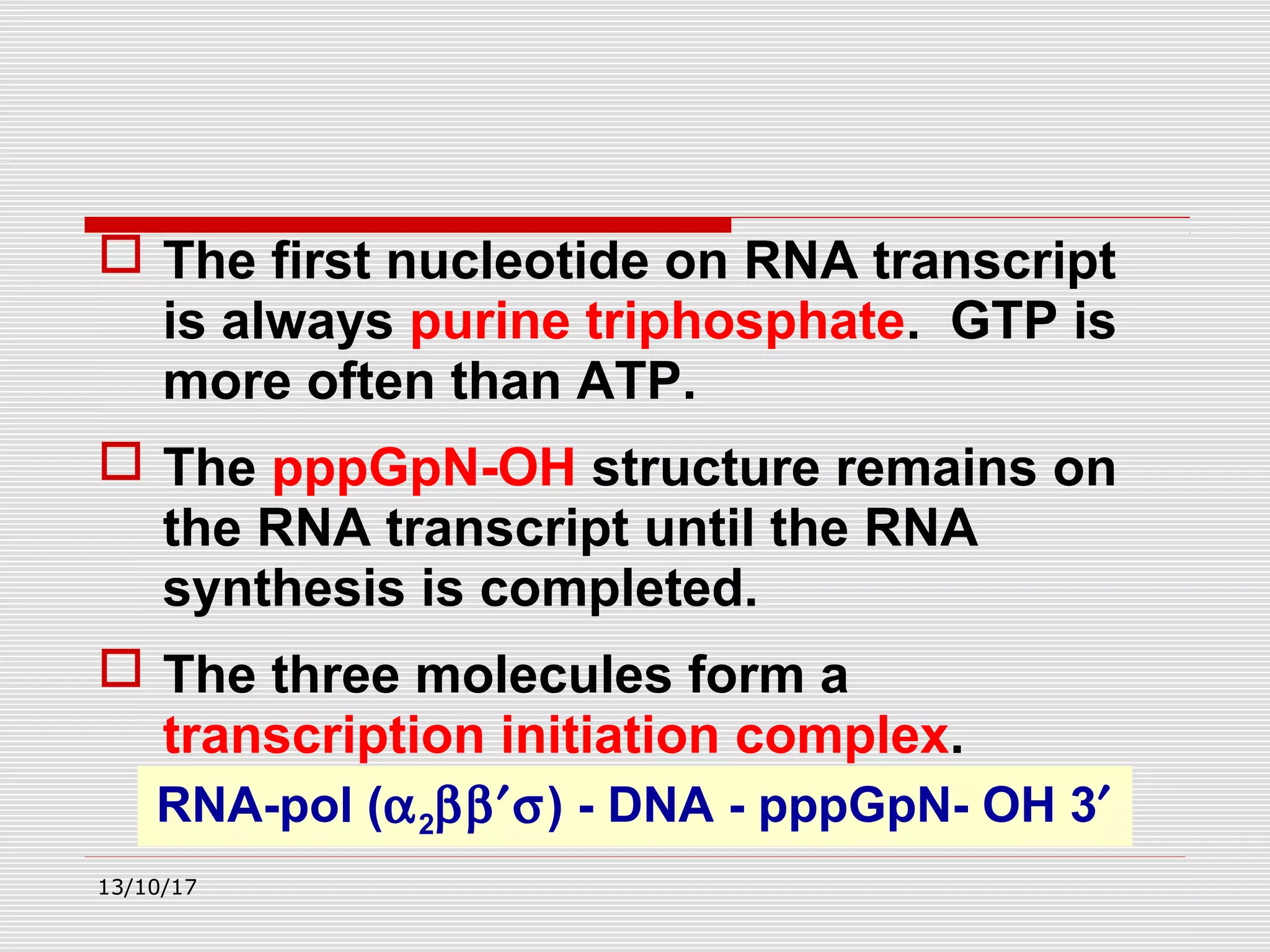
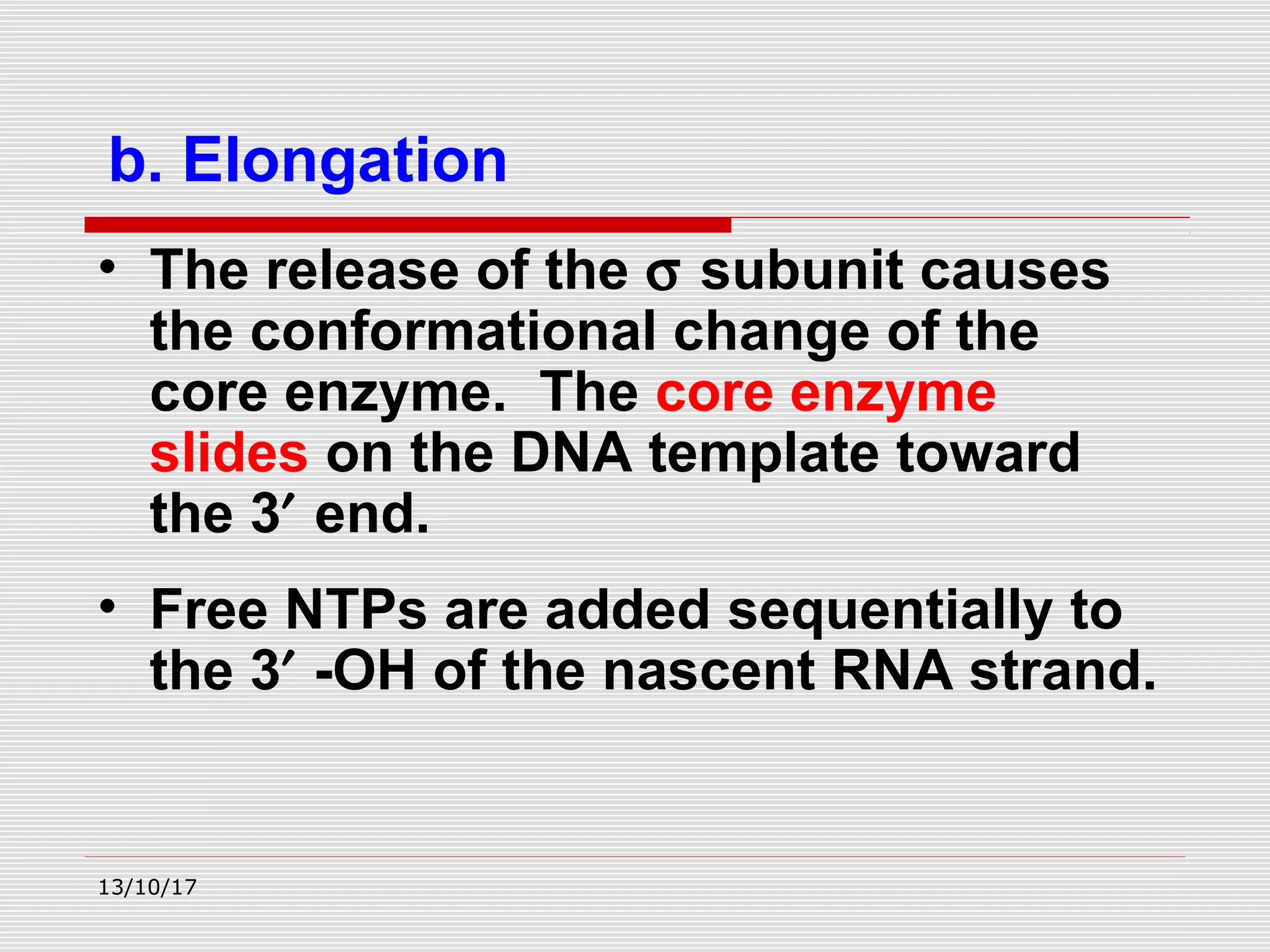
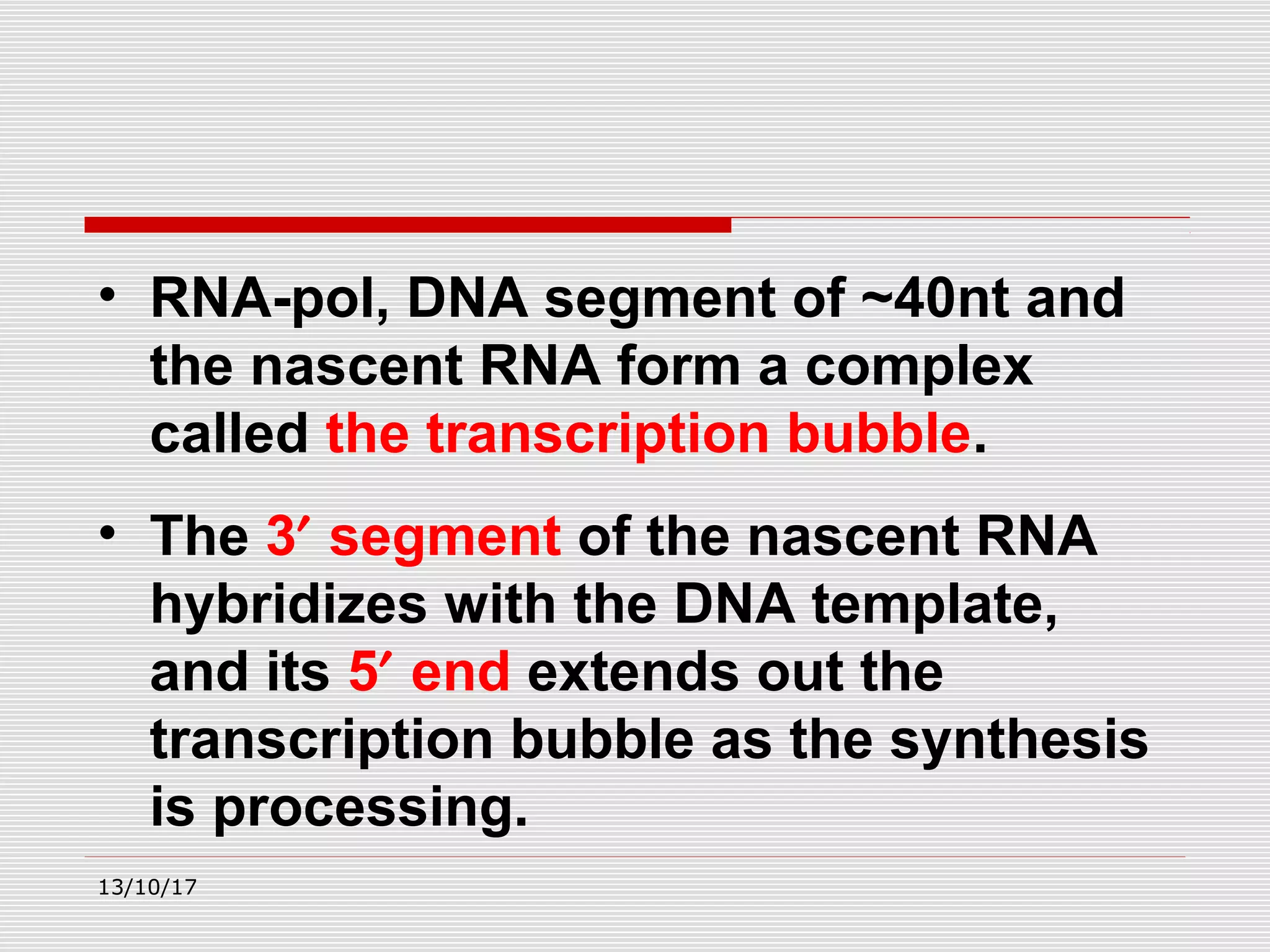
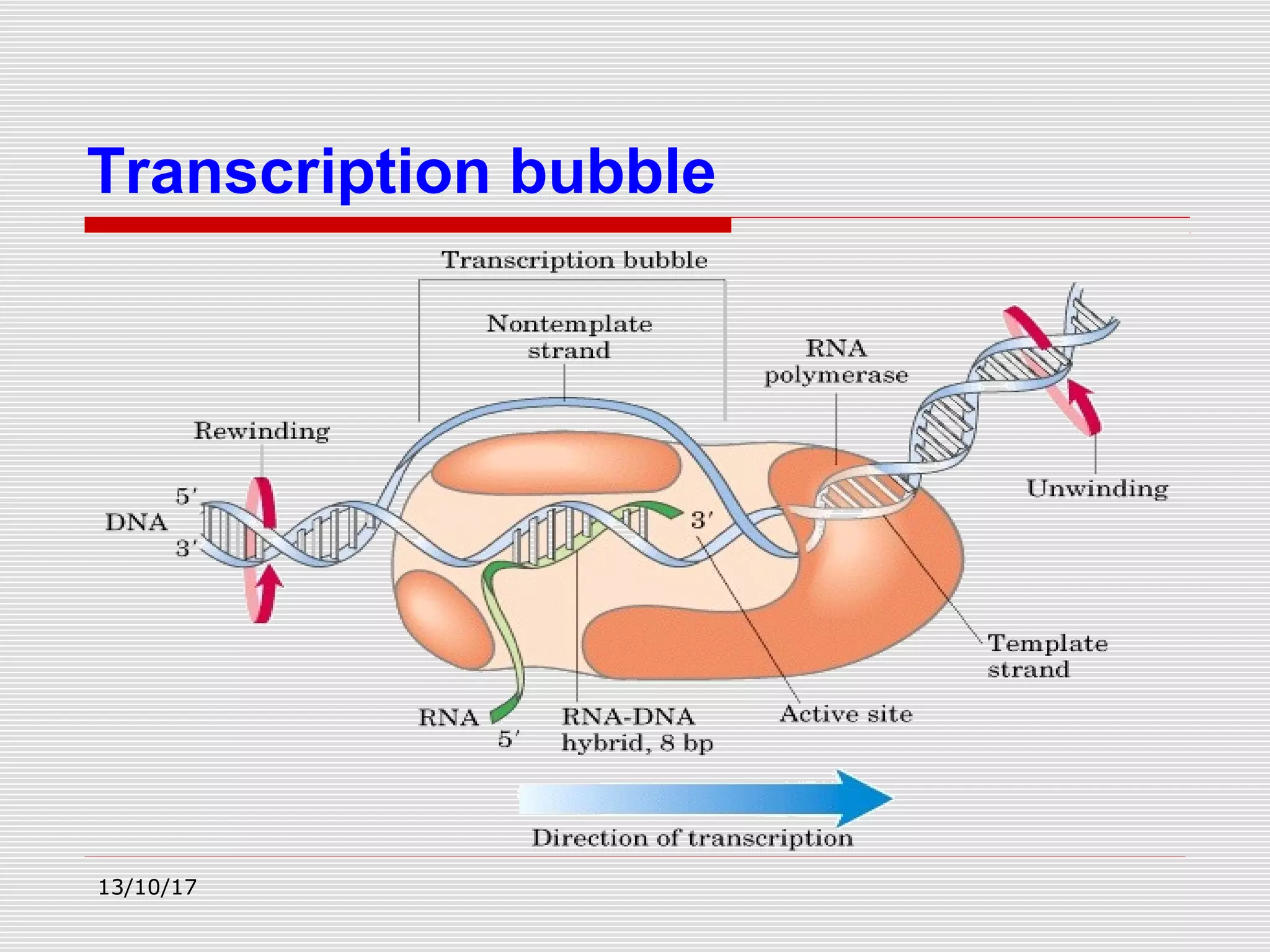
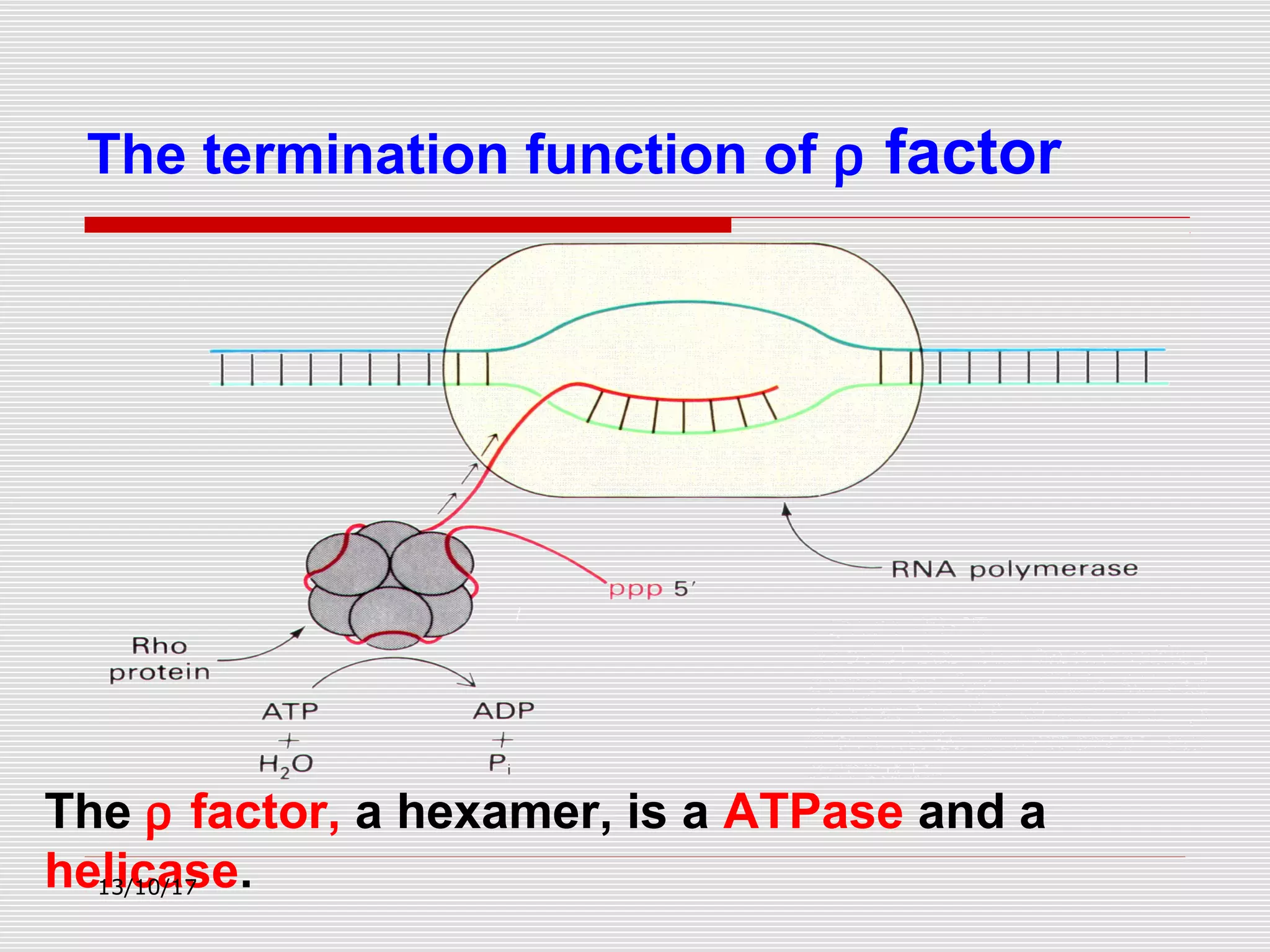
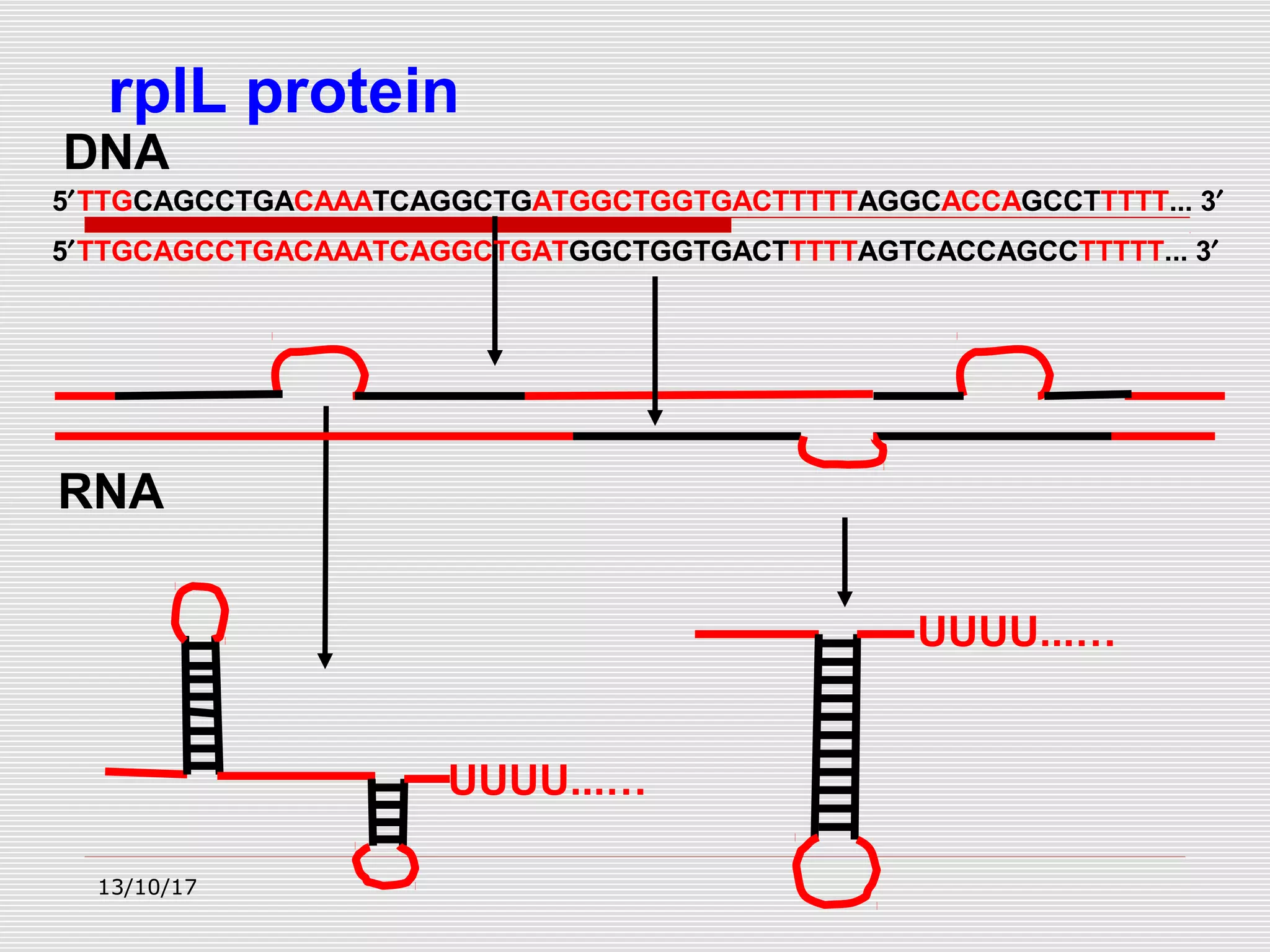
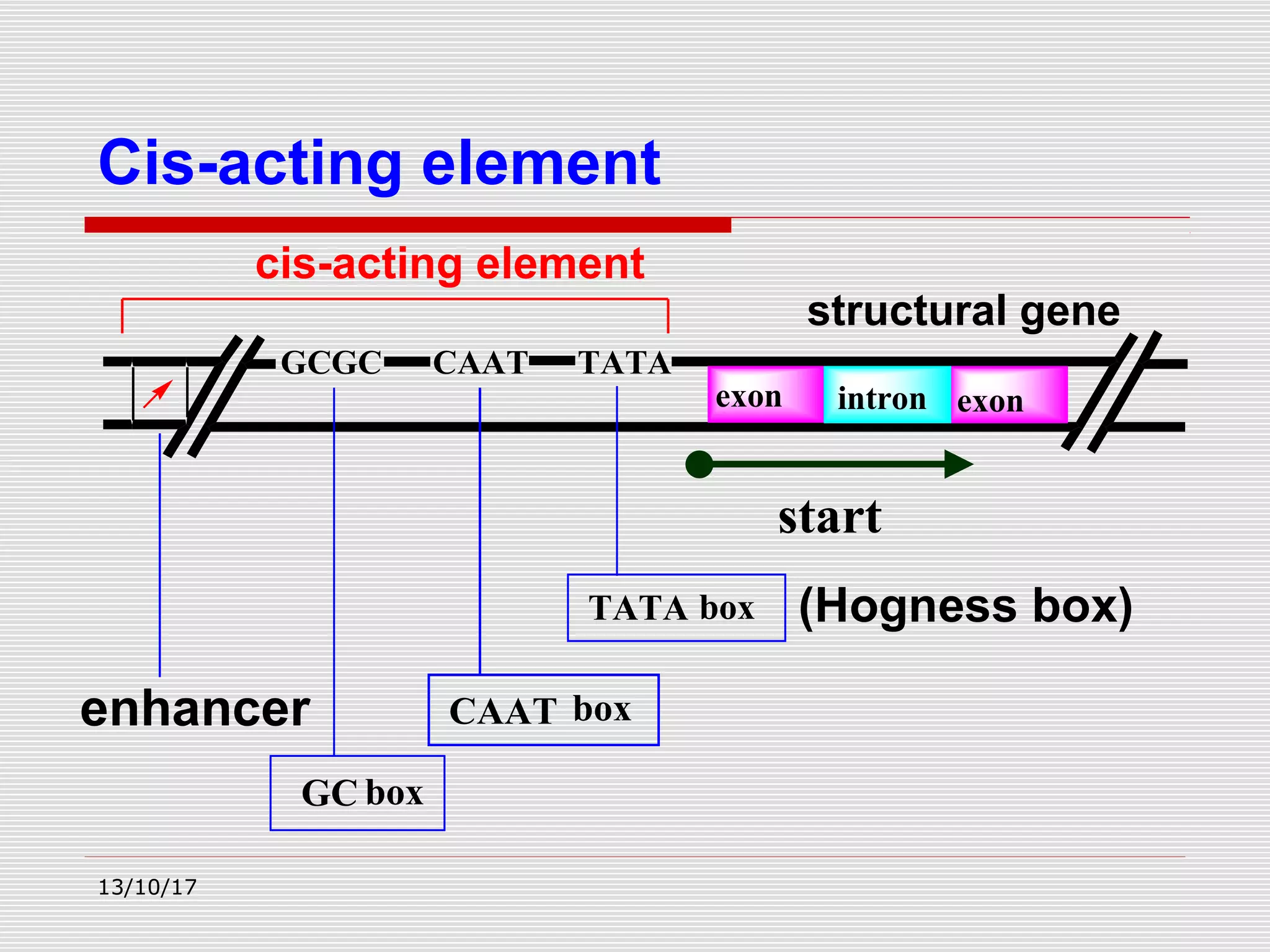
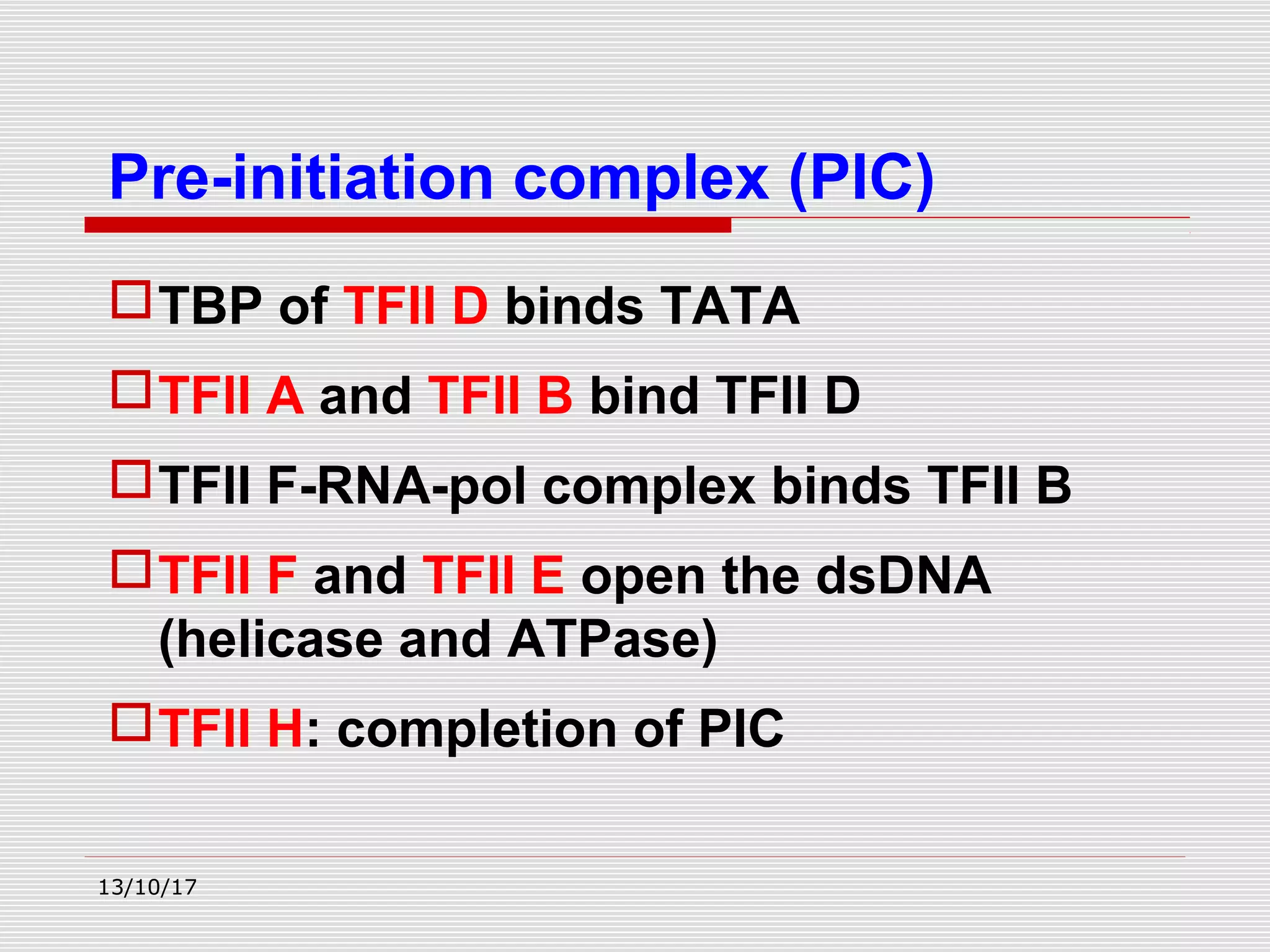
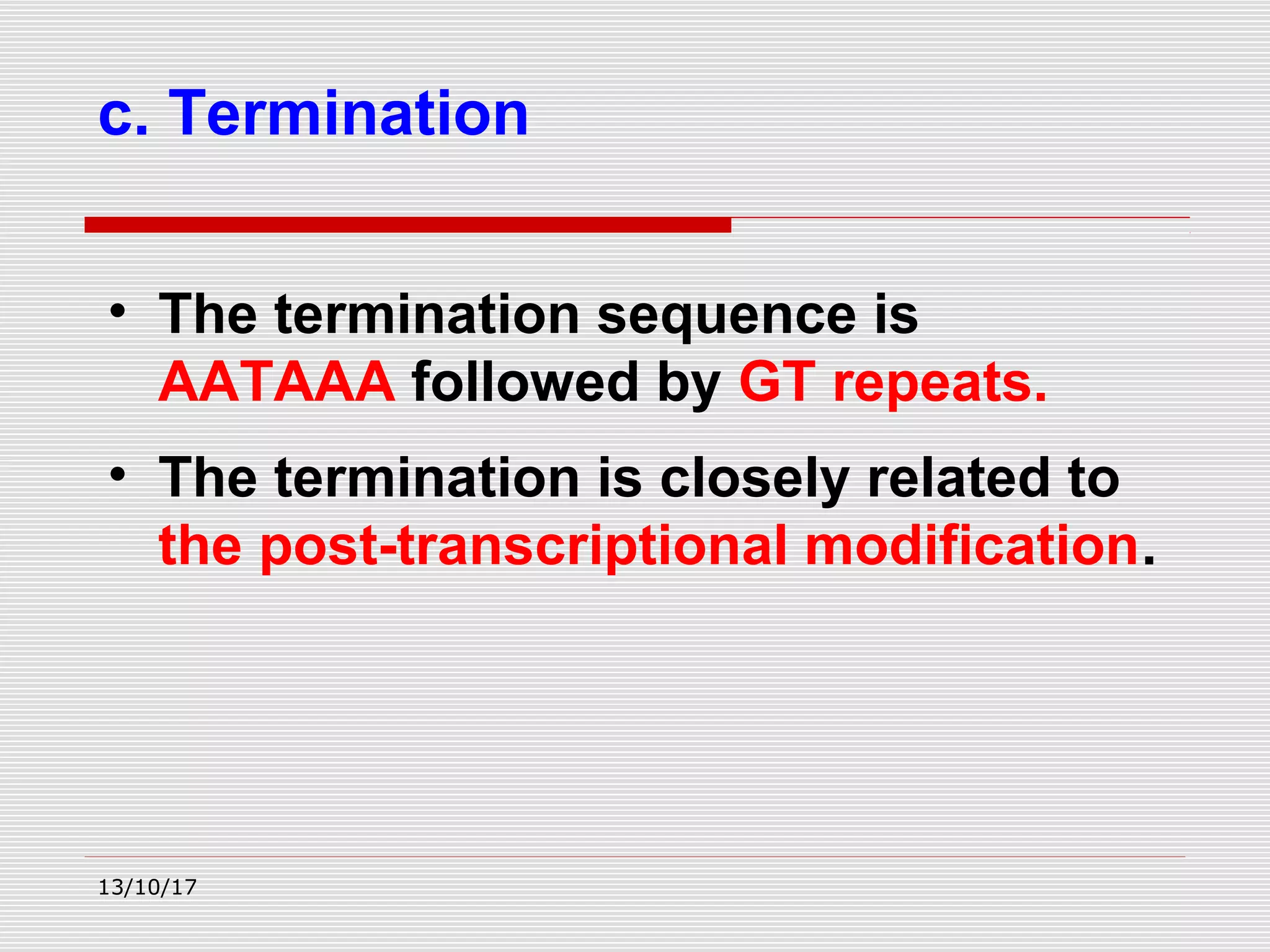
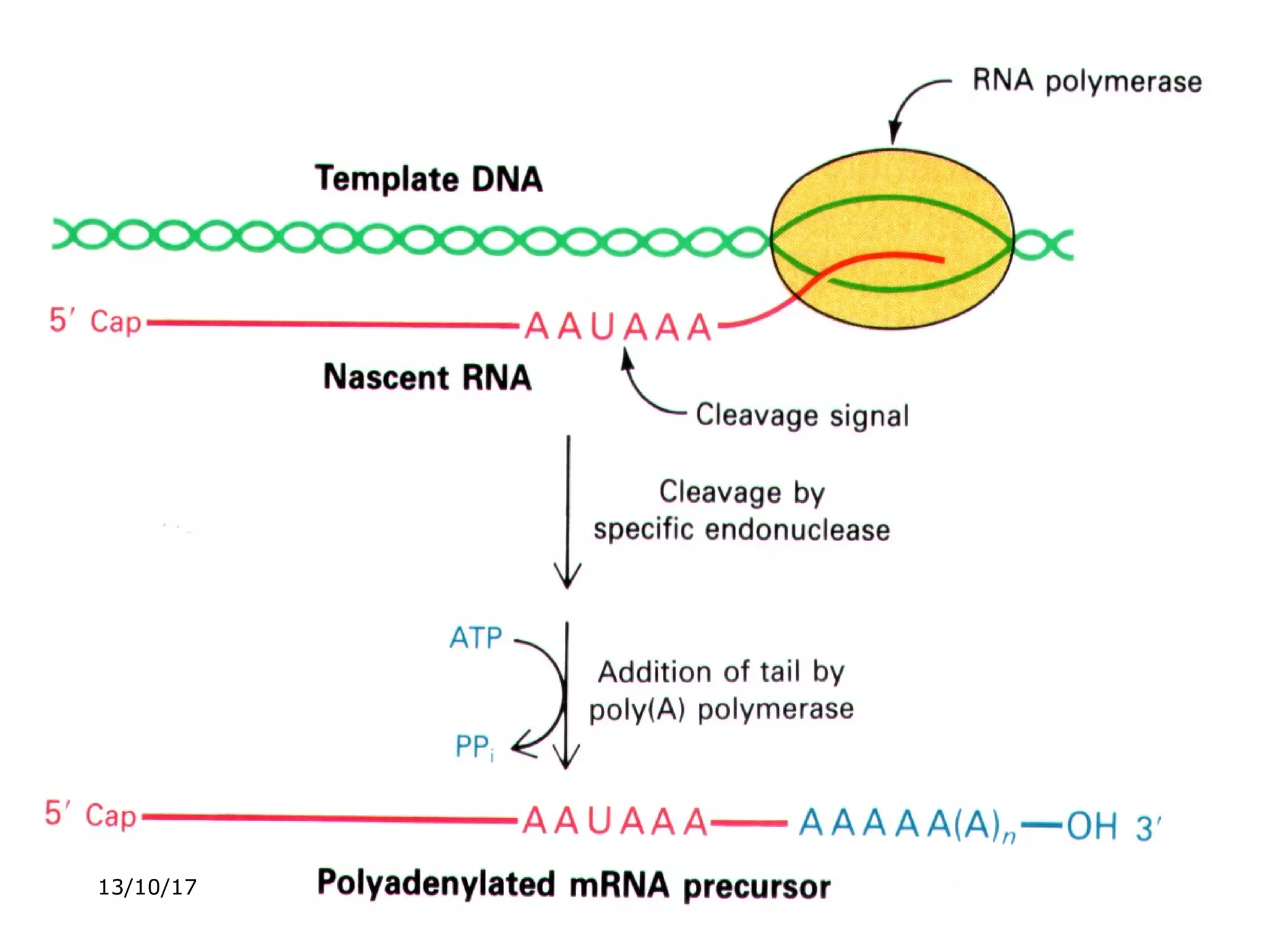
The document discusses the process of transcription in prokaryotes and eukaryotes. It describes how transcription uses DNA as a template to synthesize RNA, with RNA polymerase catalyzing the formation of phosphodiester bonds between nucleotides. The key stages of transcription - initiation, elongation, and termination - are explained for both prokaryotes and eukaryotes. Recognition sequences in DNA, such as the TATA box and transcription factors involved, differ between the two domains of life.