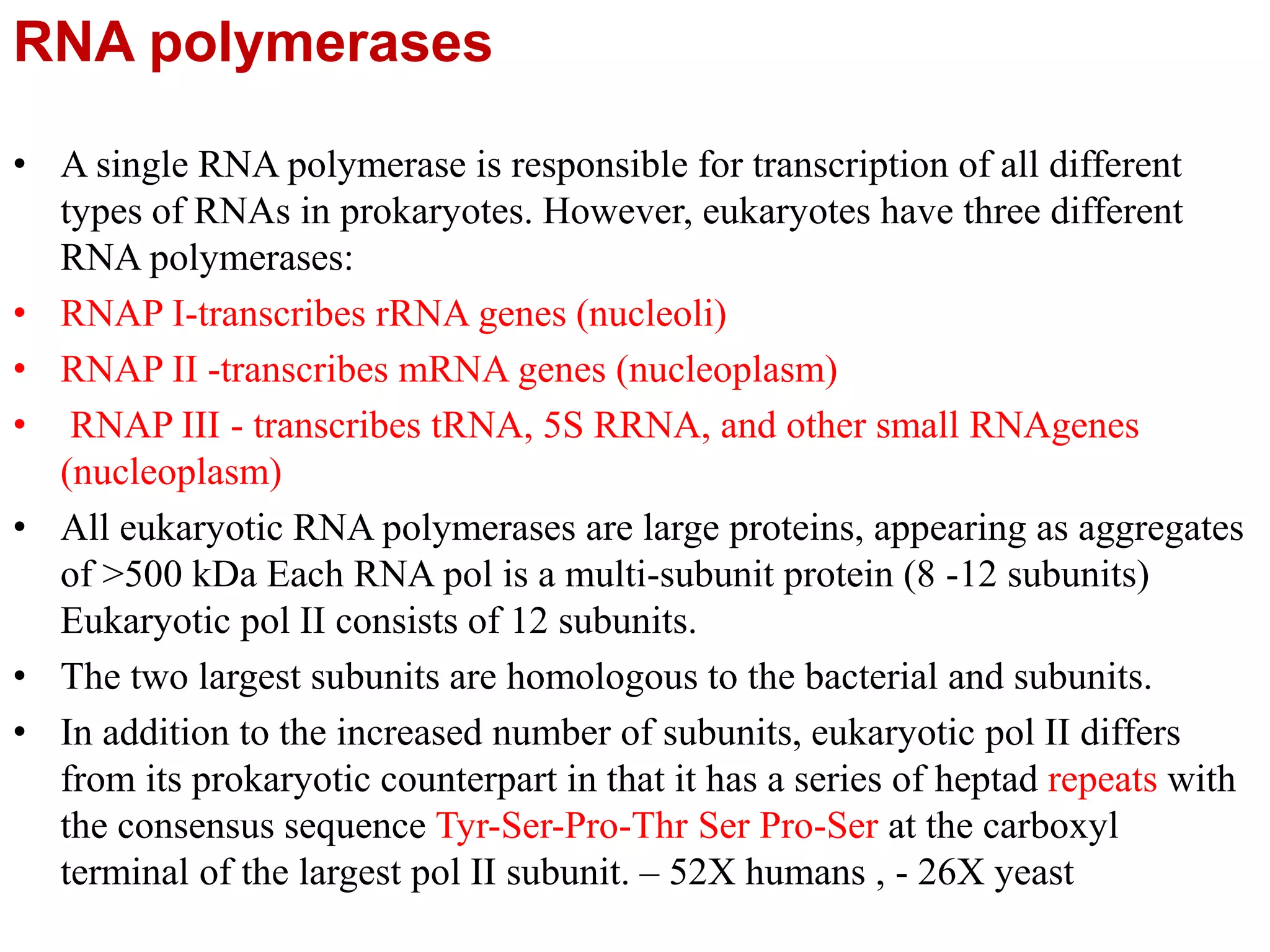
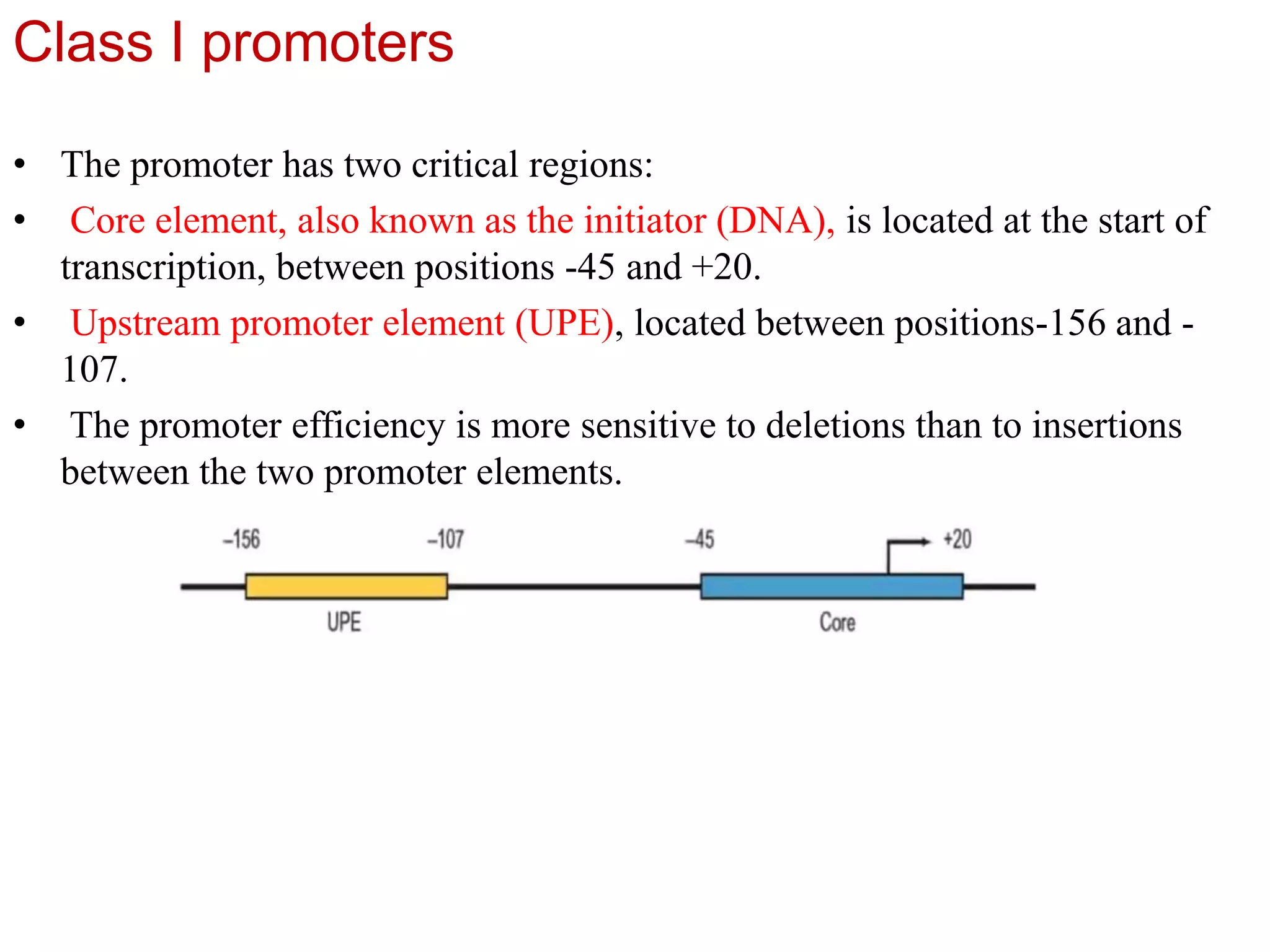
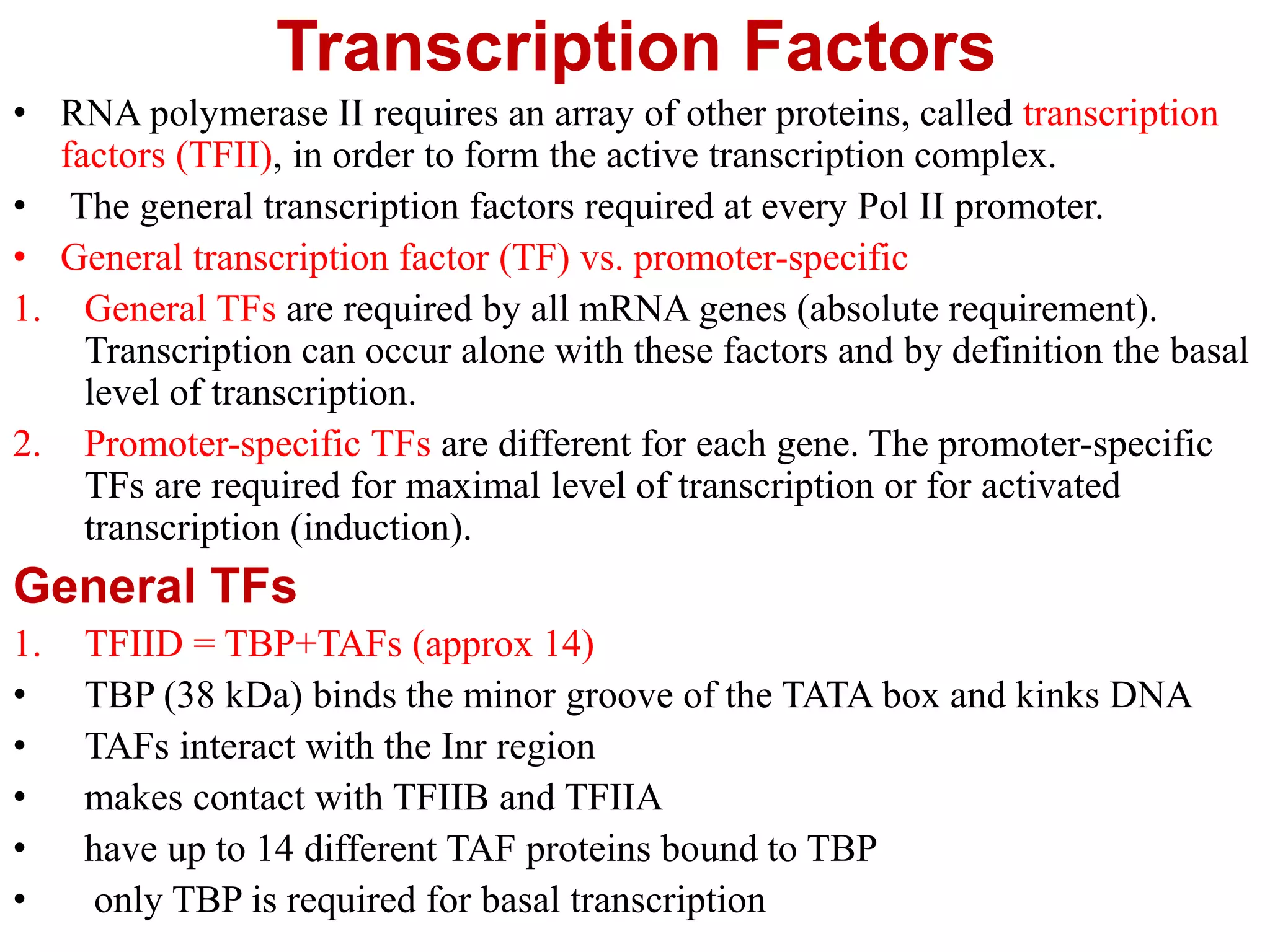
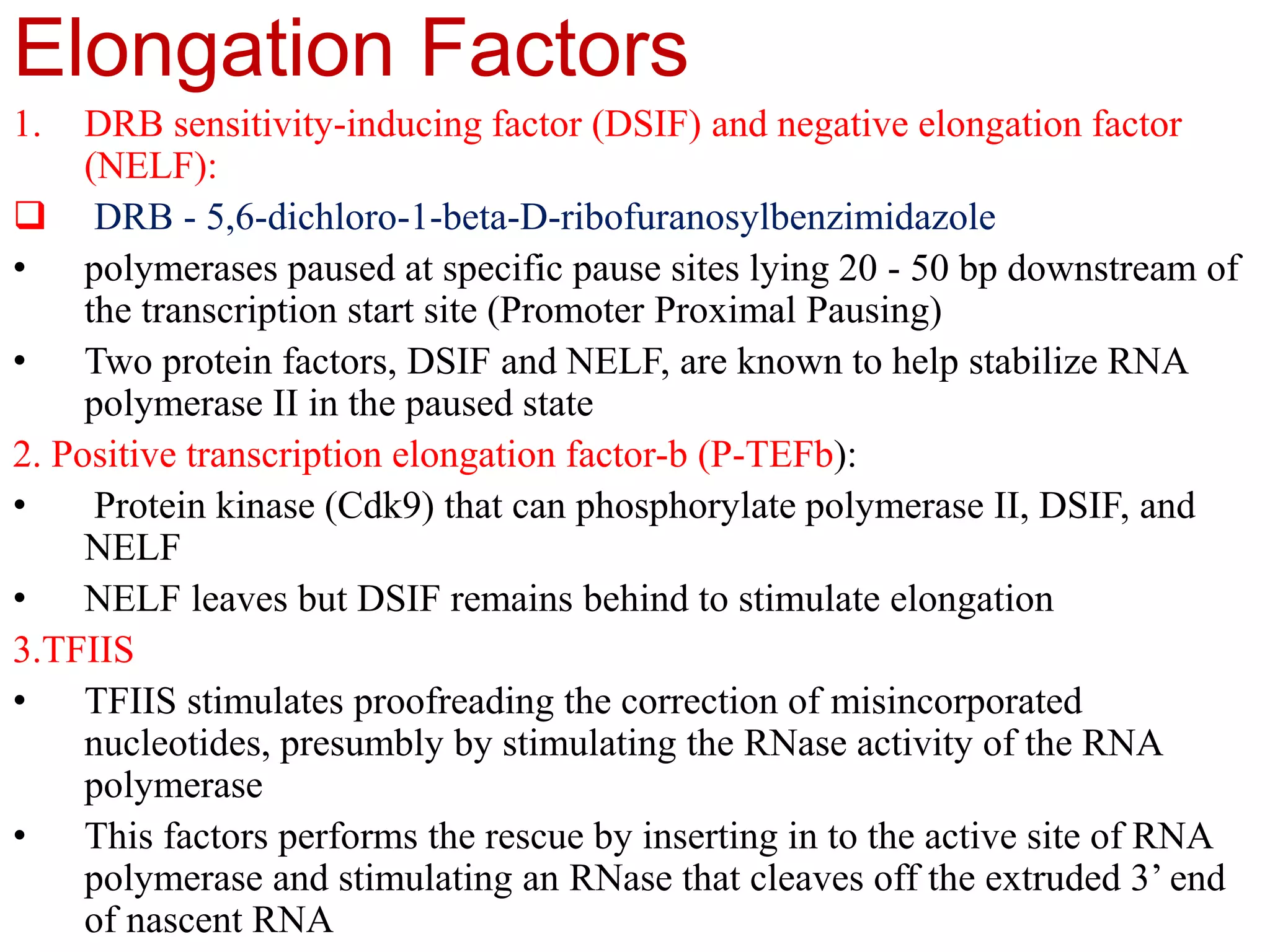
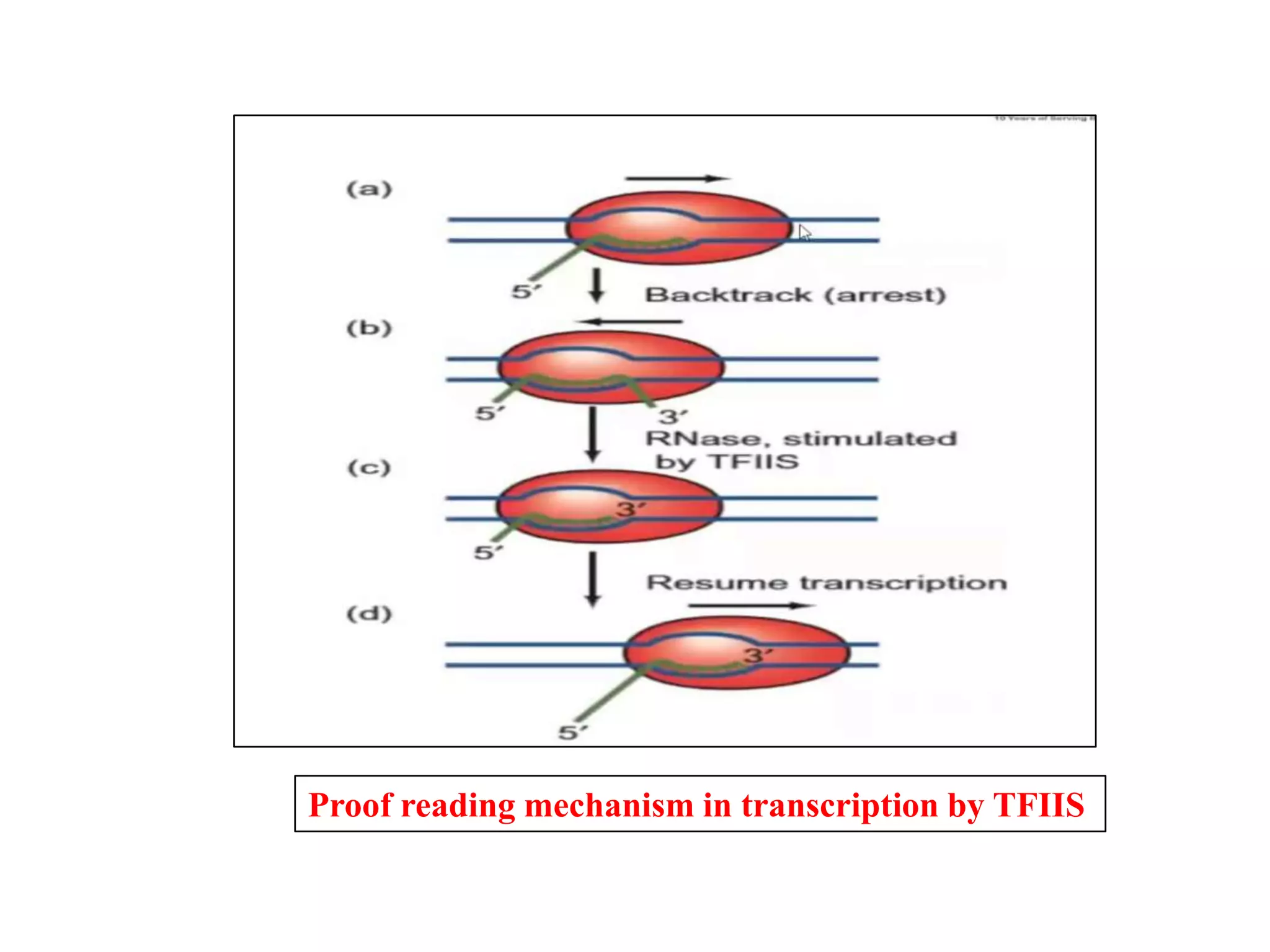
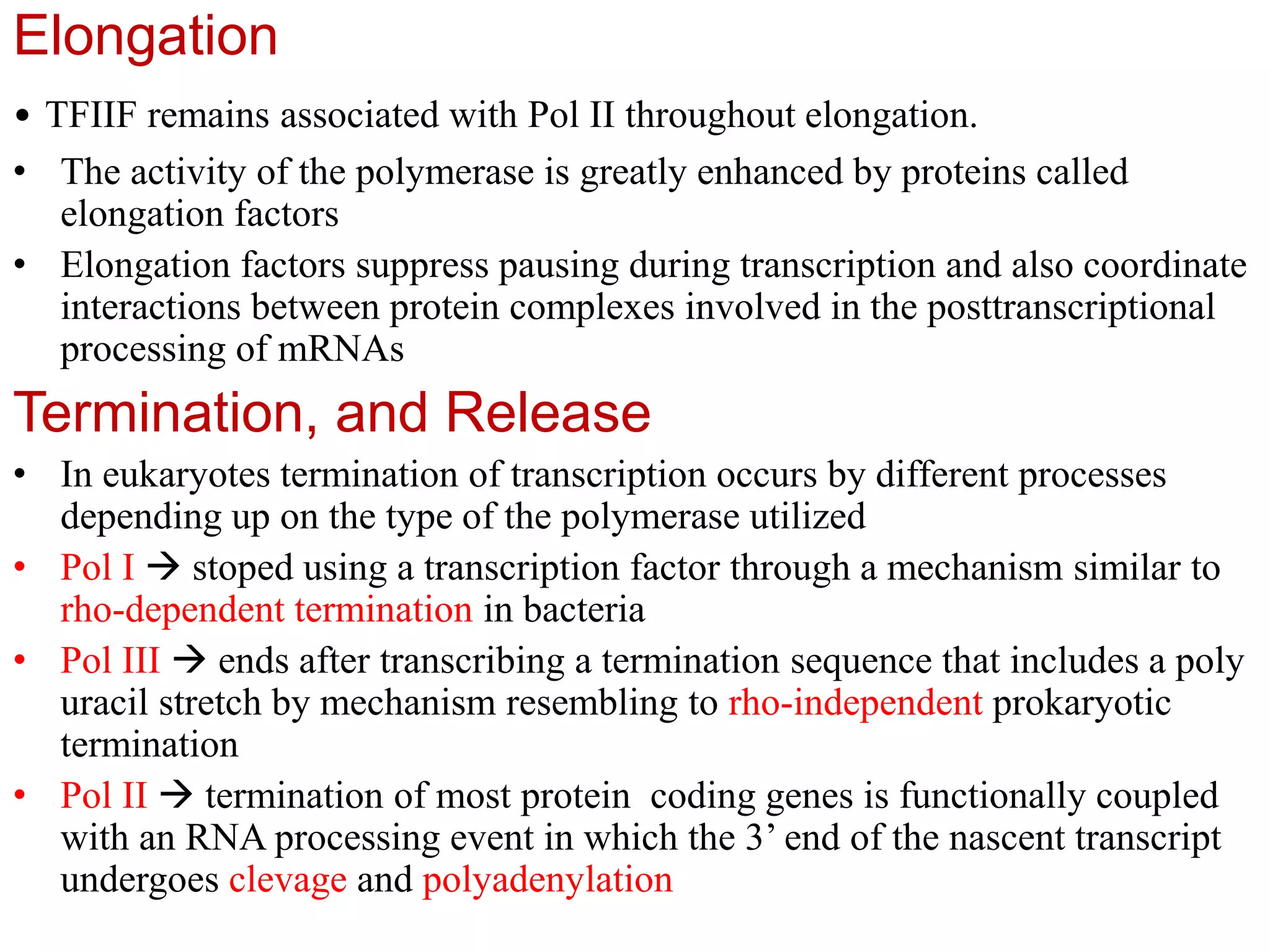
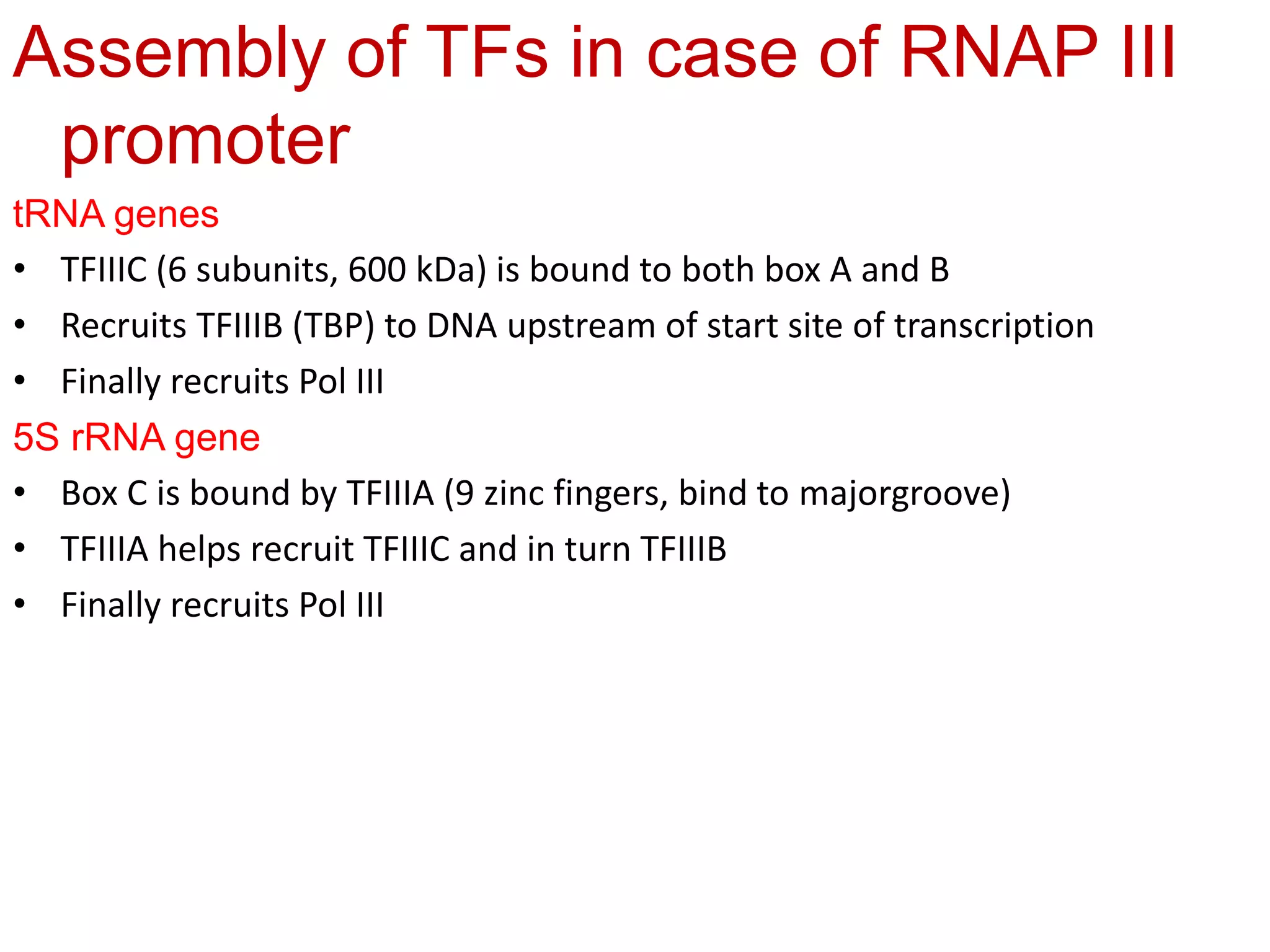
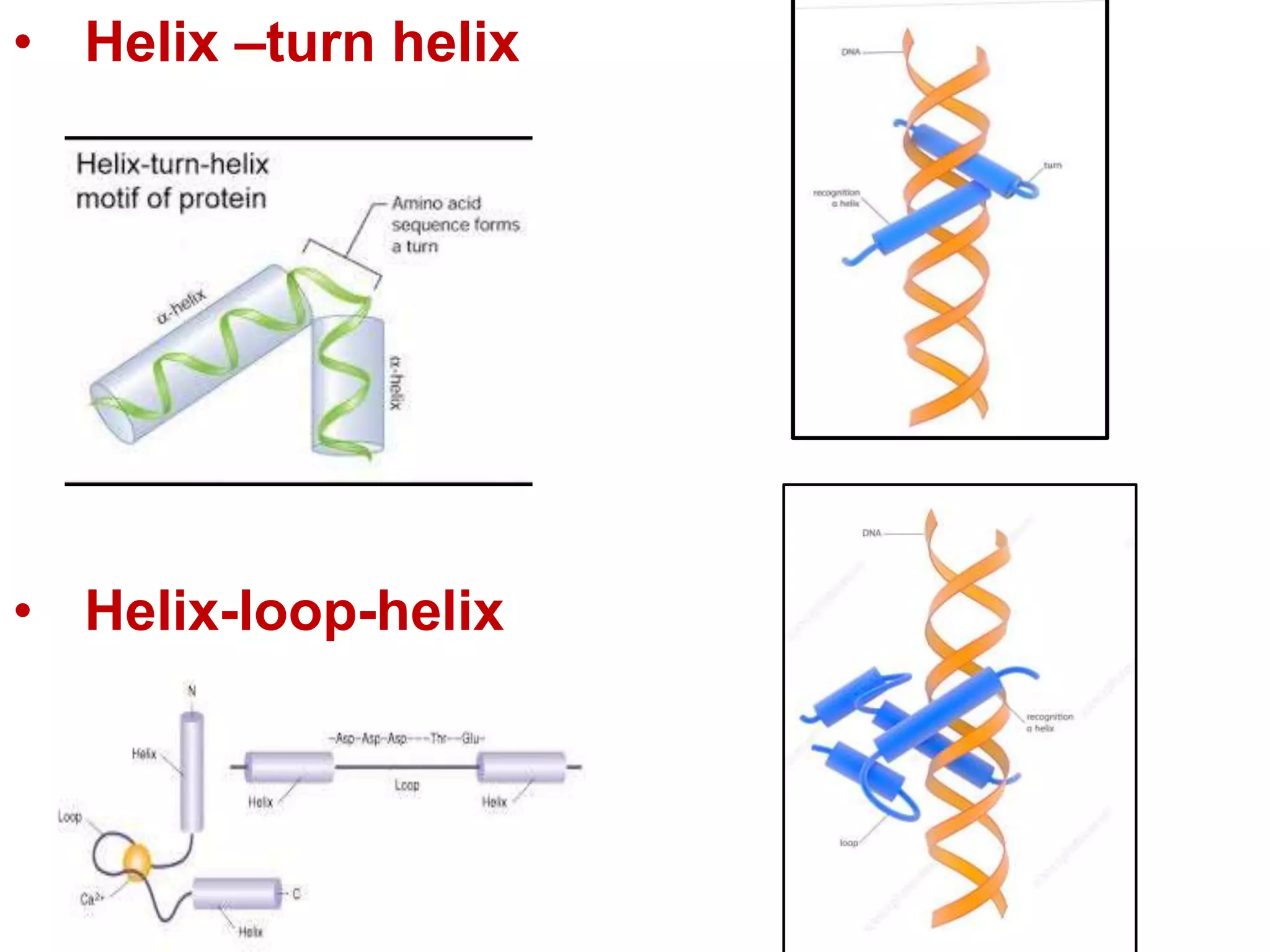
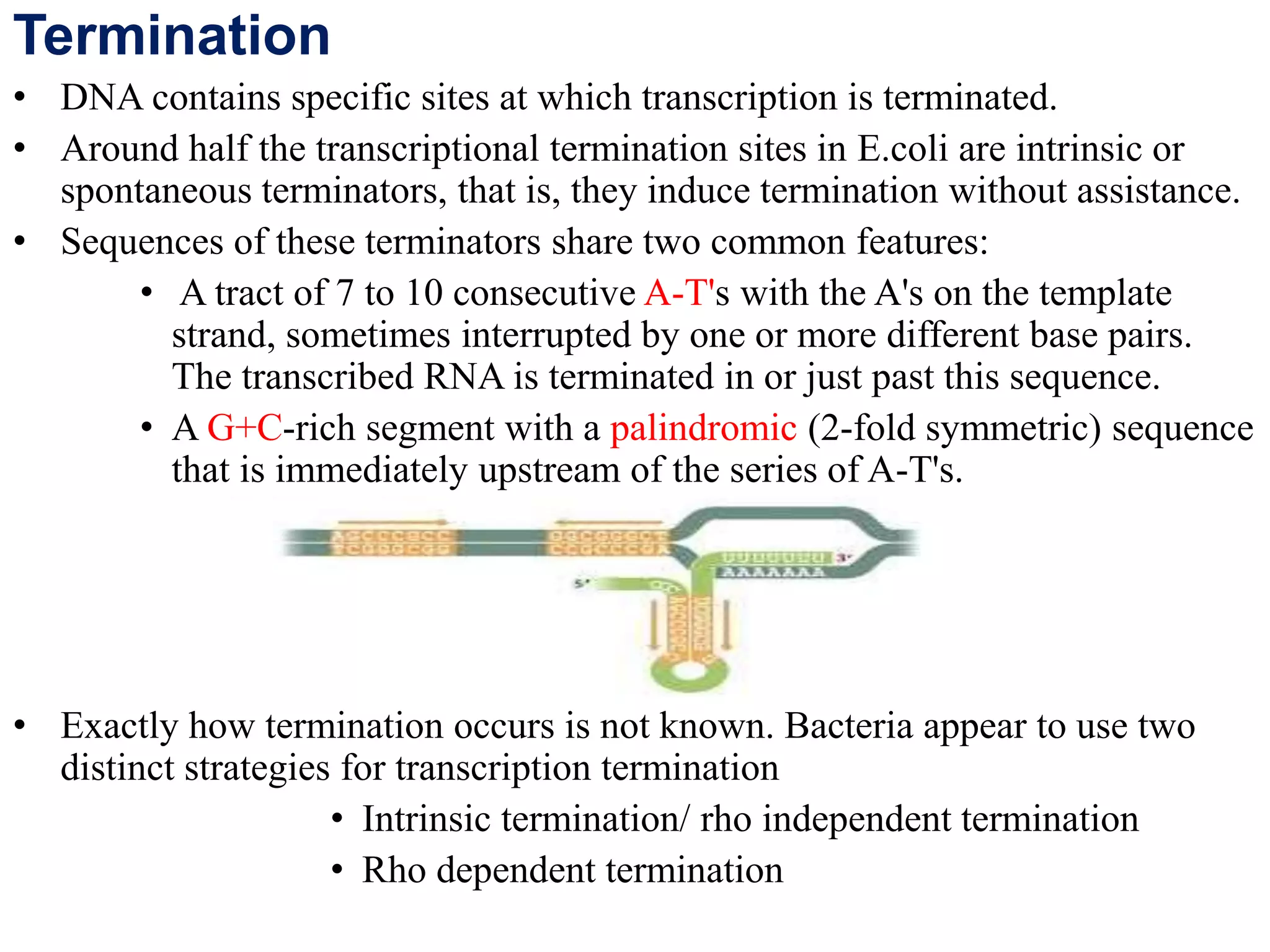
Transcription is the first step in gene expression, where RNA is synthesized from a DNA template by RNA polymerase. It involves specific regulatory sequences for initiation and termination, and results in the production of mRNA, tRNA, and rRNA. In prokaryotes, transcription is carried out by a single RNA polymerase, whereas eukaryotes have three different RNA polymerases, each responsible for transcribing different types of RNA.

























![• Rho attaches to nascent RNA at its recognition sequence [named rut (for
Rho utilization), a C-rich segment of at least 40 nt]Translocates along the
RNA in the 5' 3' direction until it encounters an RNAP paused at the
termination site
• Rho pushes the RNAP forward in a way that partially rewinds its dsDNA
helix at the transcription bubble while unwinding the RNA-DNA hybrid
helix thus releasing the RNA.](https://image.slidesharecdn.com/transcription-200827060841/75/Transcription-30-2048.jpg)