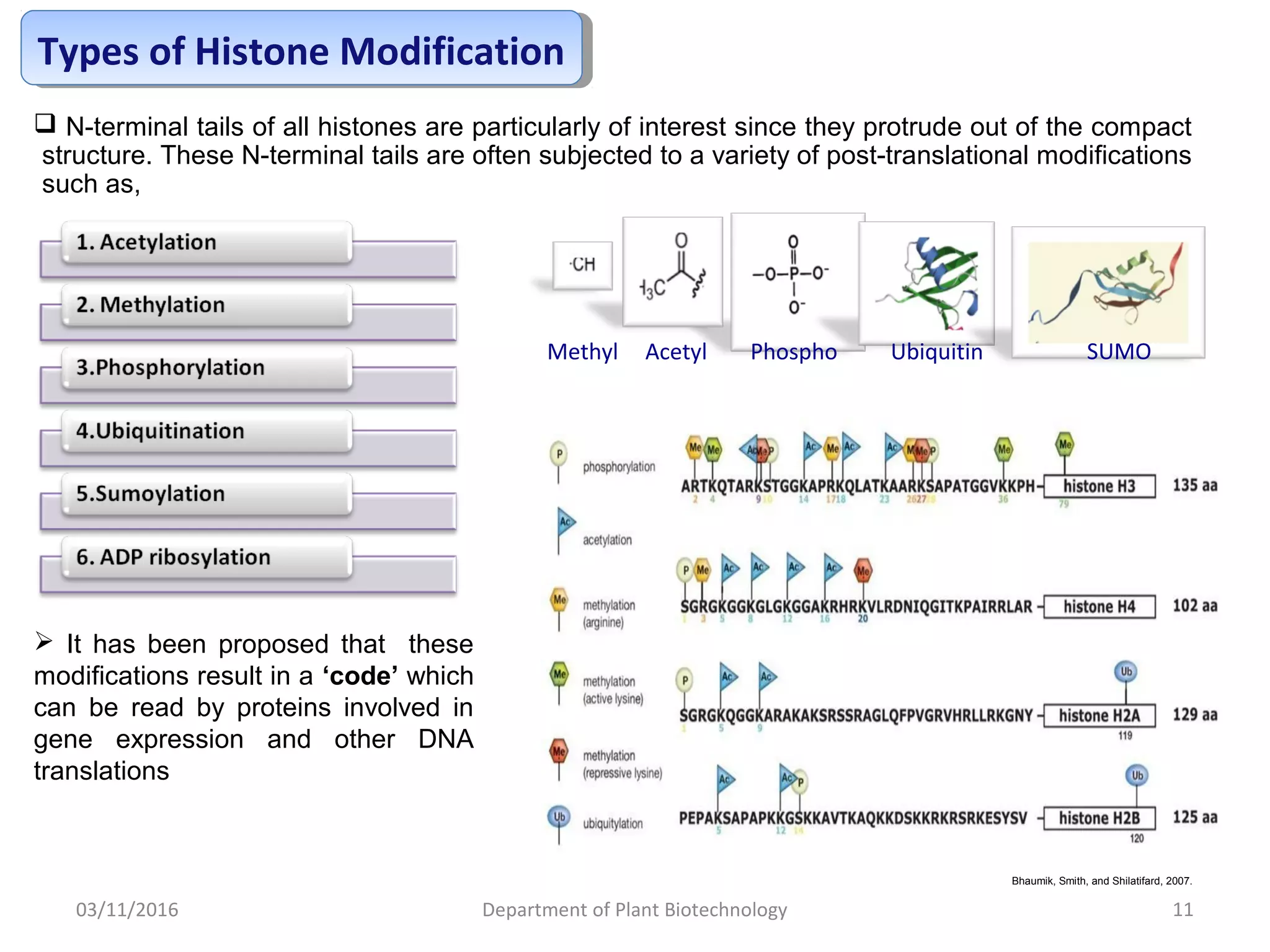
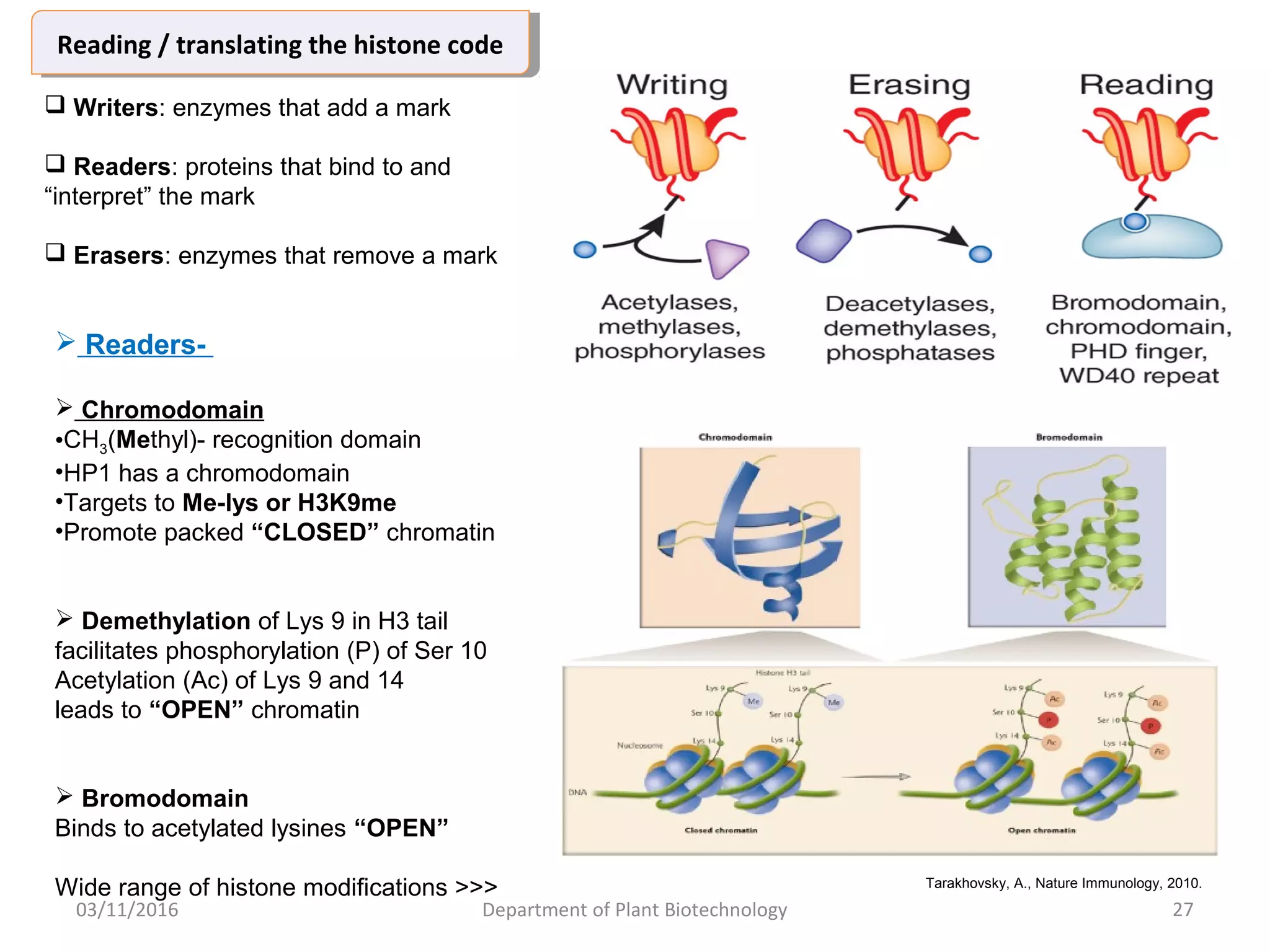
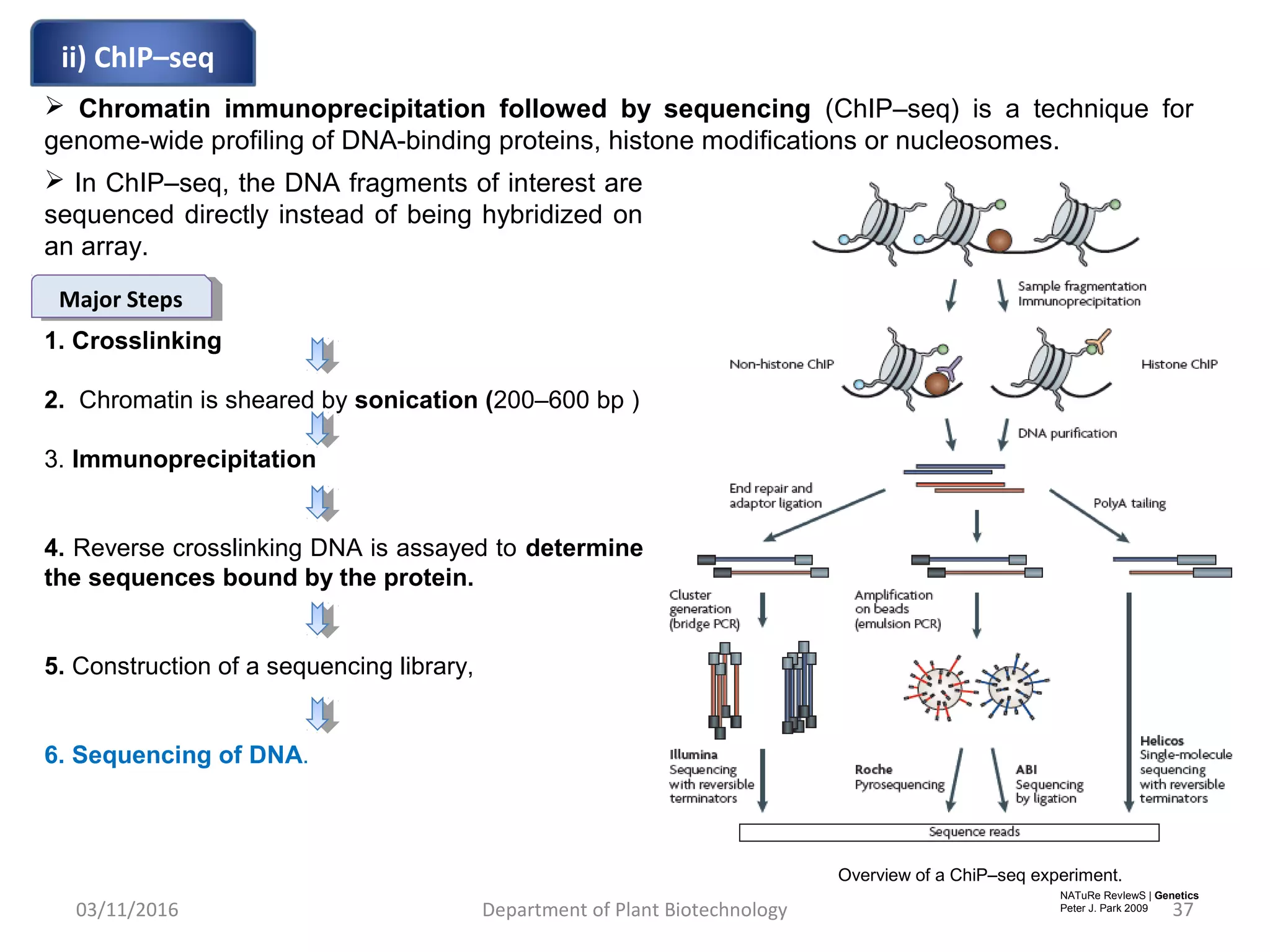
The document discusses histones, essential proteins that package DNA into chromatin, highlighting their structure, types, and role in gene regulation. It details various post-translational modifications (PTMs) of histones, including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and adp-ribosylation, which significantly impact chromatin structure and gene expression. The interactions and modifications of histones serve as a dynamic mechanism for regulating gene activity and cellular functions.