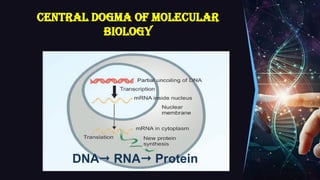
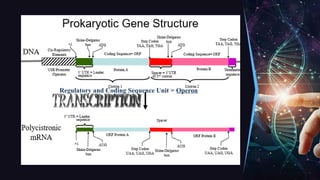
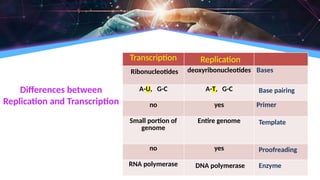
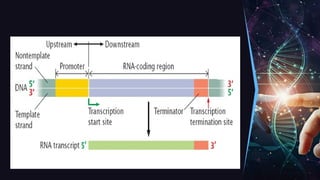
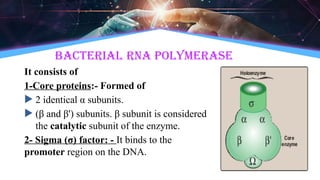
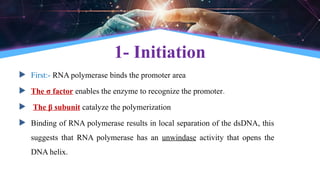
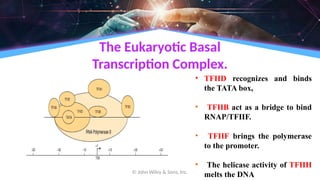
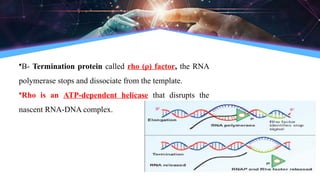
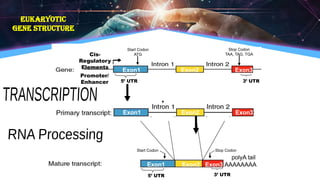
The document outlines the process of transcription, detailing the roles of prokaryotic and eukaryotic promoters, RNA polymerases, and the steps involved in RNA synthesis, including initiation, elongation, and termination. It explains the structural differences between RNA types and the components involved in the transcription unit. Additionally, it highlights the distinctions between transcription and replication, along with the specific requirements for RNA synthesis.