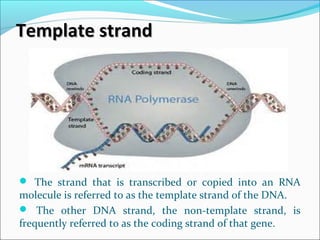
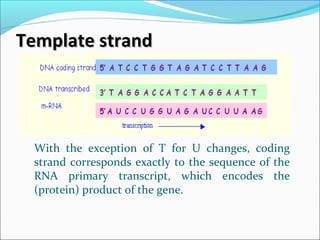
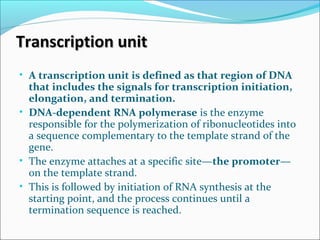
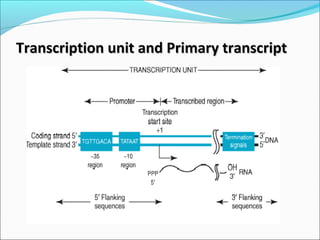
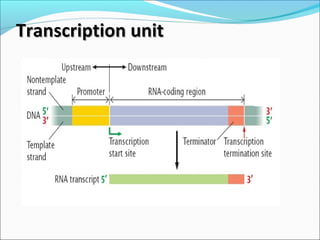
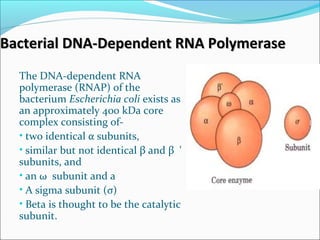
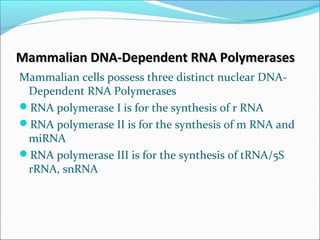
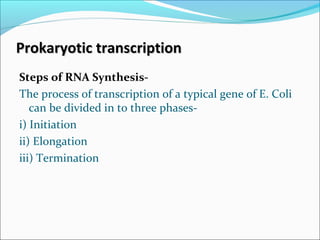
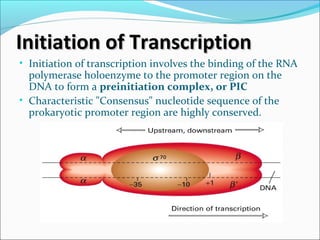
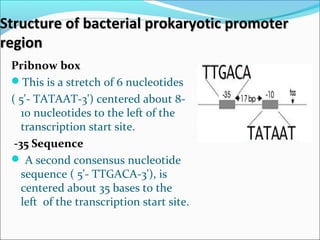
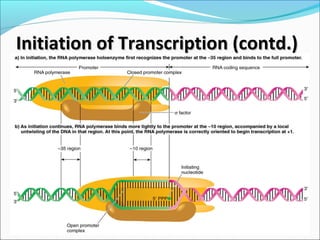
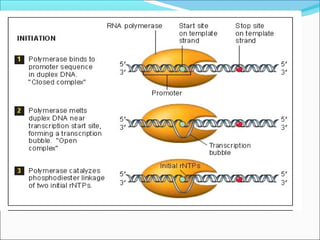
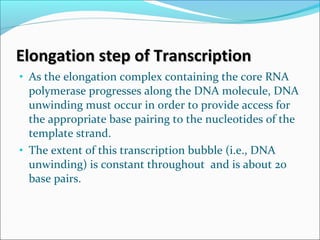
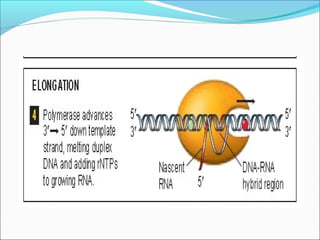
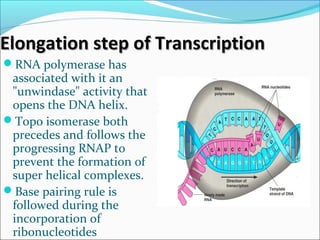
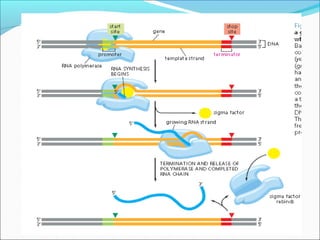
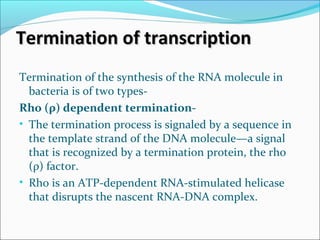
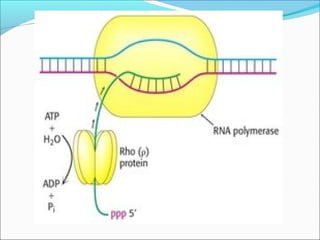
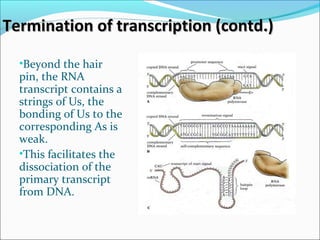
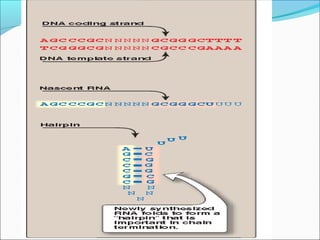
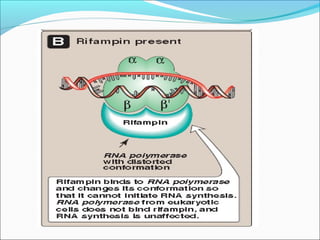
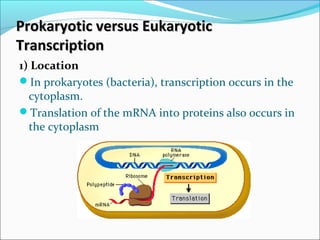
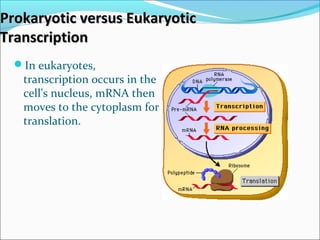
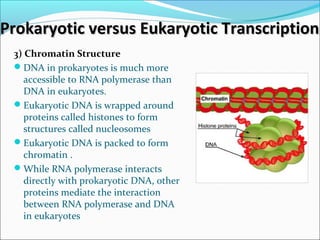
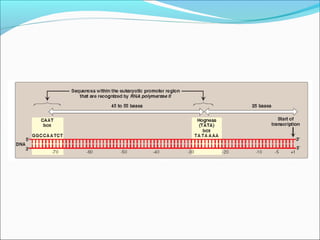
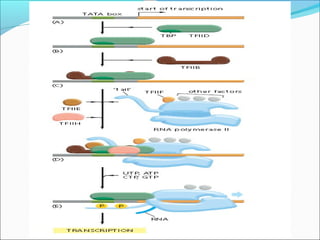
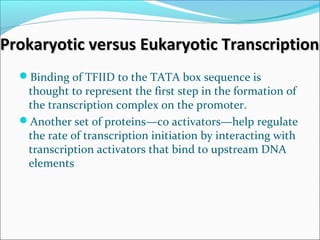
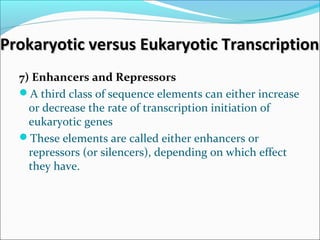
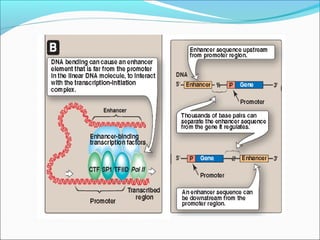
The document discusses DNA transcription in prokaryotes and eukaryotes. It defines transcription as the synthesis of RNA from a DNA template. In eukaryotes, transcription occurs in the nucleus and involves RNA polymerases I, II, and III. The process includes initiation, elongation, and termination. Eukaryotic transcription is more complex than prokaryotic transcription due to larger genome size, chromatin structure, and presence of regulatory elements. The primary transcript in eukaryotes undergoes processing before becoming a mature mRNA.