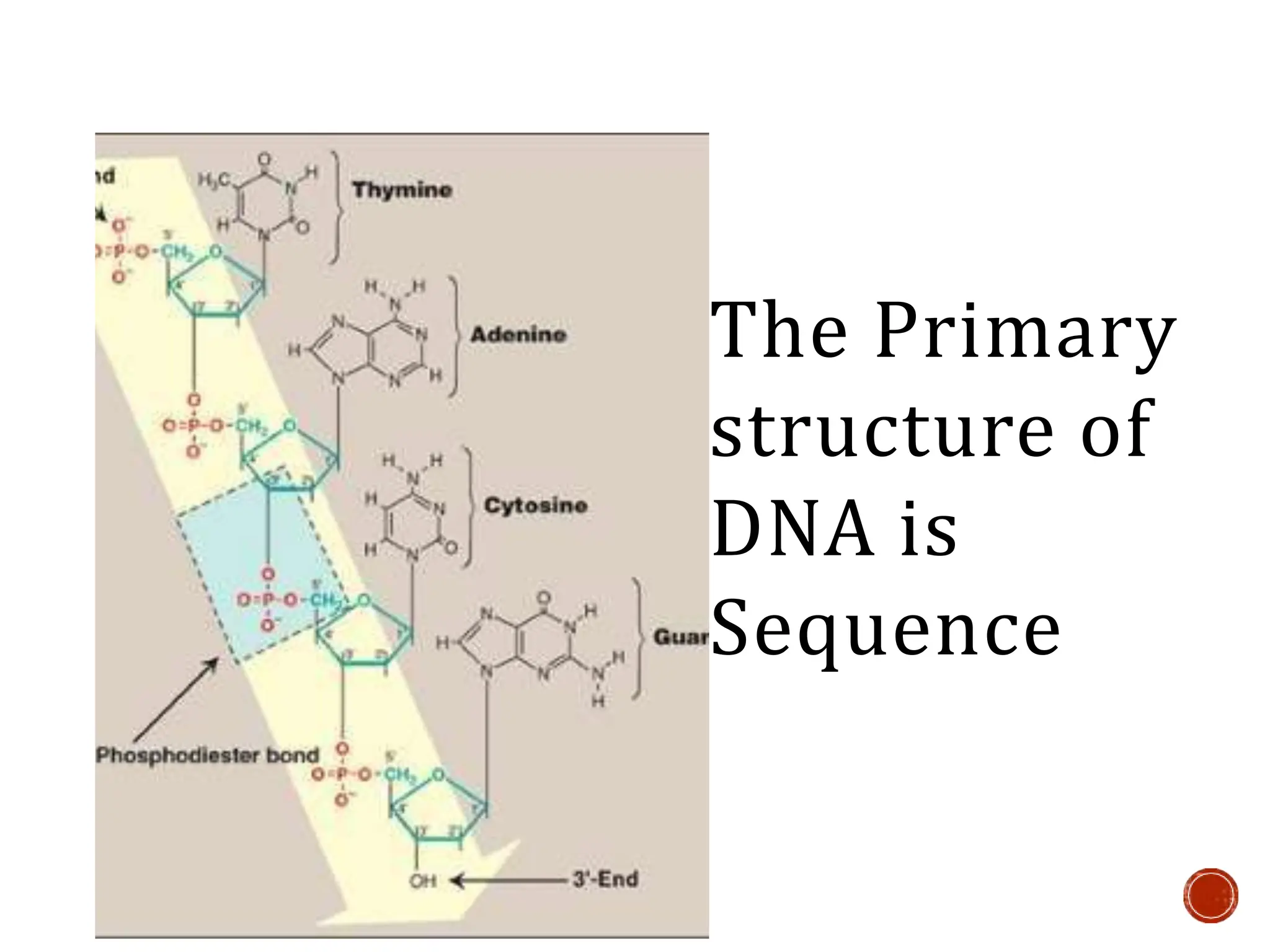
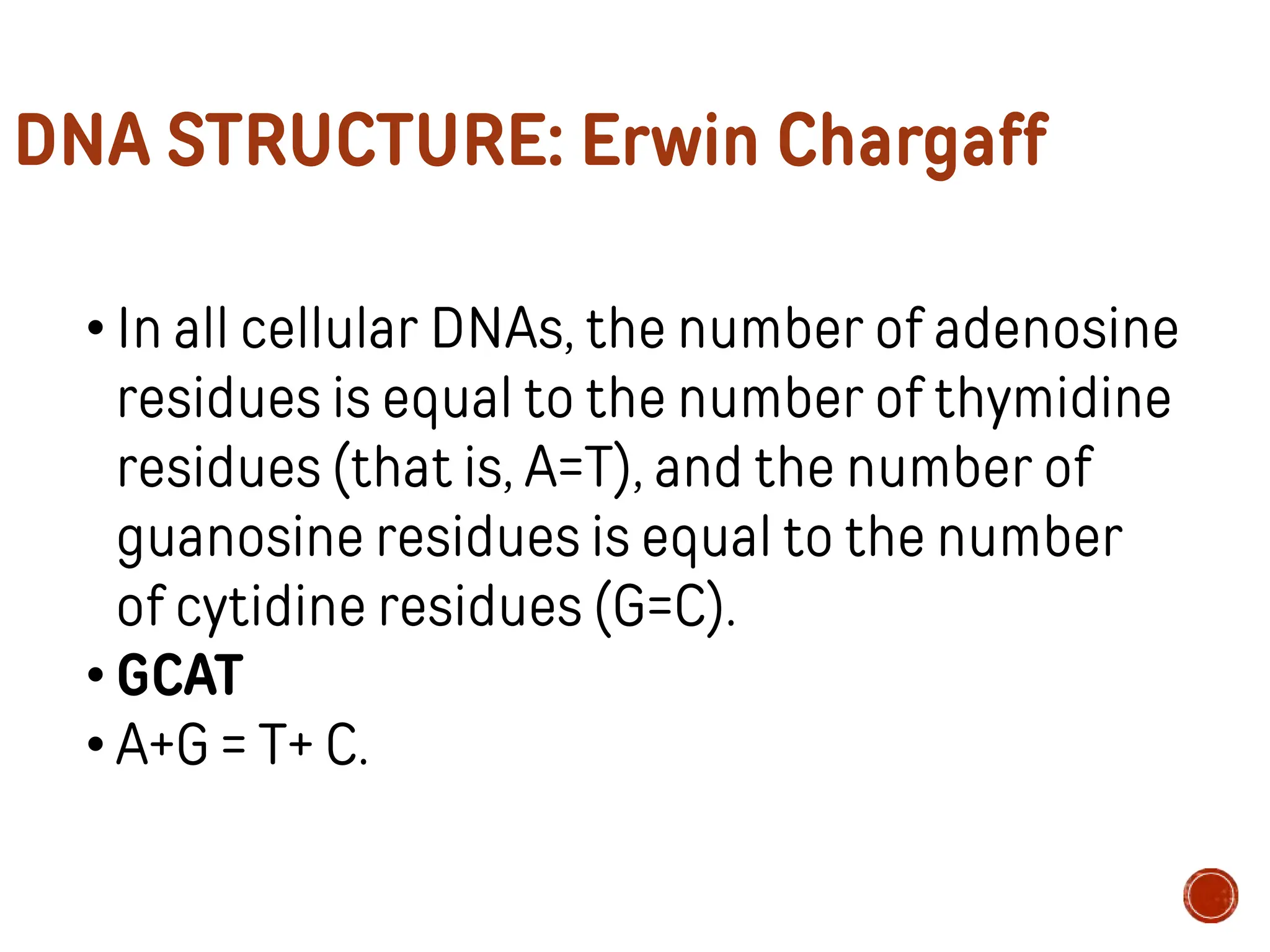
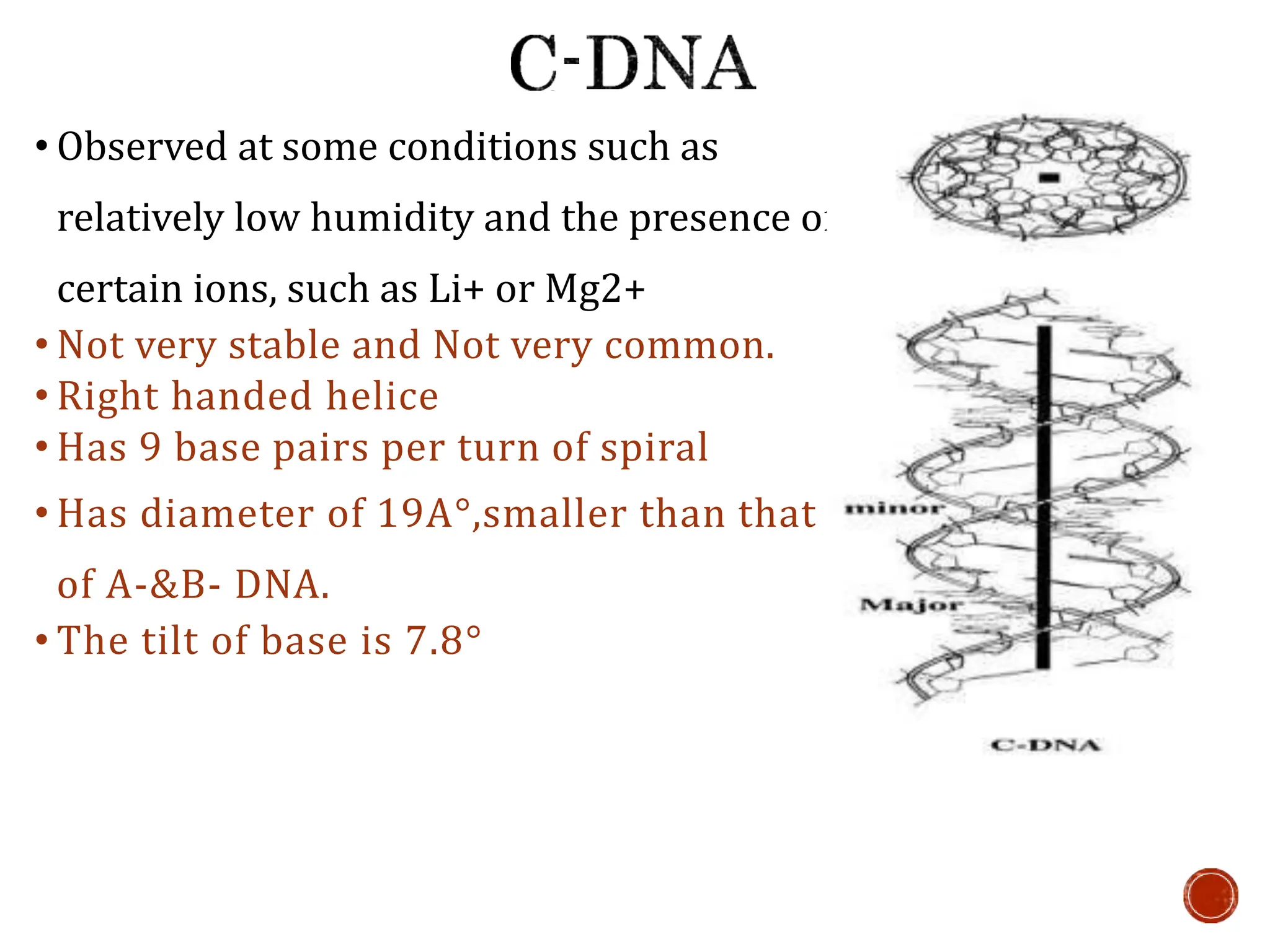
This document provides information about DNA structure and types. It begins with a timeline of important discoveries in DNA research. It then discusses the primary and secondary structures of DNA, including the double helix model proposed by Watson and Crick. It describes Chargaff's rules and the complementary base pairing of A-T and G-C. Finally, it summarizes the different forms of DNA like A, B, and Z-DNA and discusses mitochondrial DNA and unusual DNA sequences.