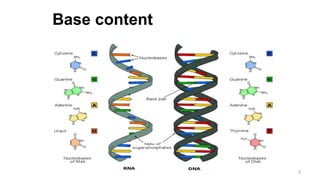
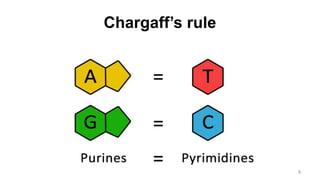
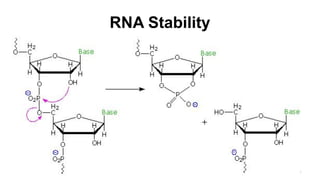
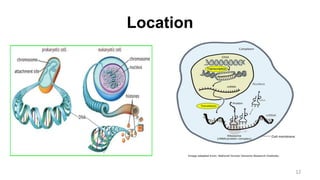
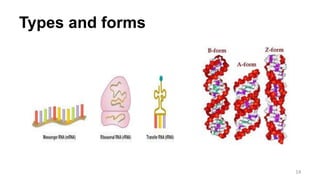
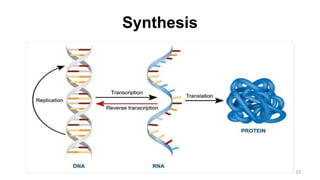
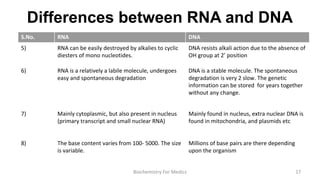
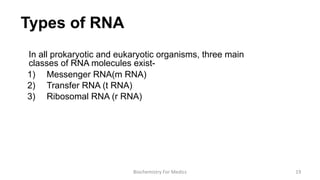
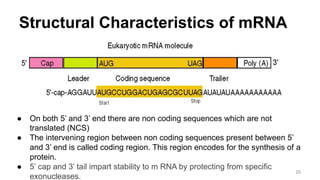
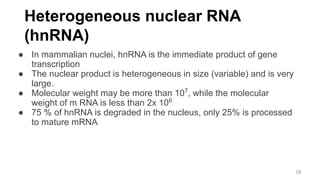
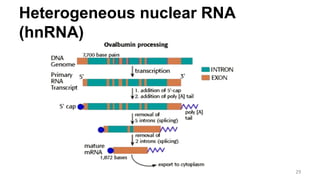
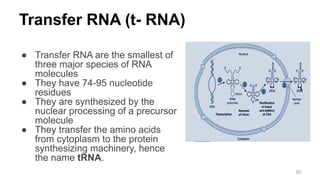
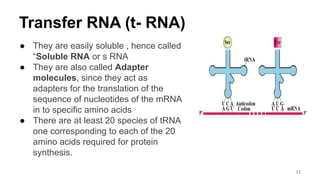
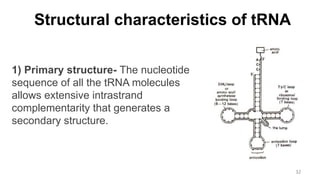
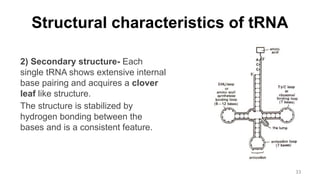
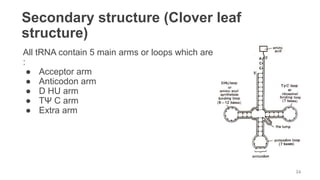
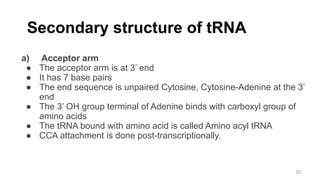
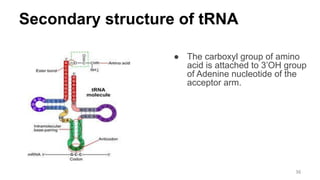
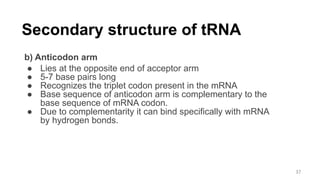
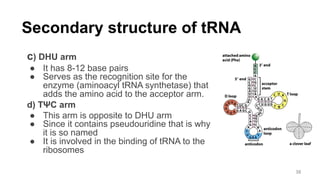
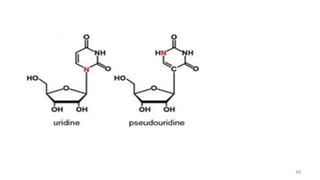
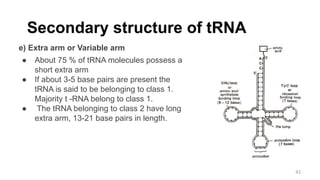
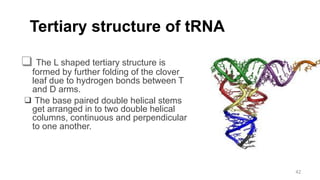
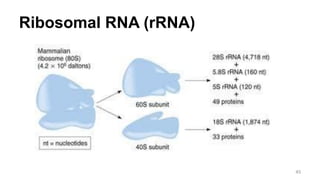
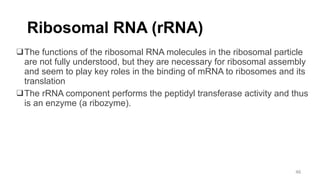
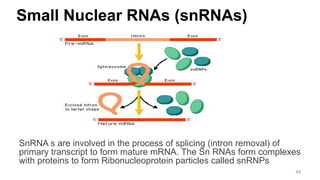
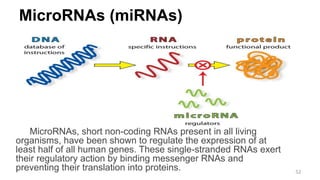
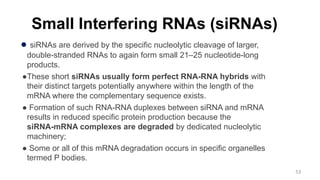
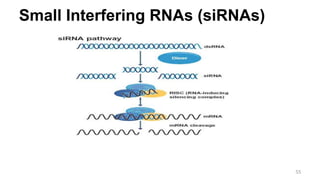
RNA exists in several forms that are involved in protein synthesis and regulation. Major RNA types include messenger RNA (mRNA), which carries genetic code from DNA to ribosomes for protein production. Ribosomal RNA (rRNA) is a core component of ribosomes where protein synthesis occurs. Transfer RNA (tRNA) transports amino acids to the ribosome during protein assembly. Small nuclear RNAs (snRNAs) help process mRNA, while microRNAs (miRNAs) and small interfering RNAs (siRNAs) regulate gene expression. Certain RNA molecules function as catalysts in cells. New research indicates miRNAs and siRNAs could be targeted for therapeutic drug development.