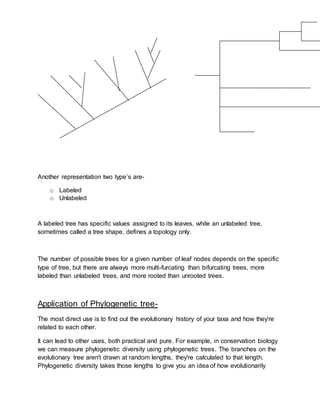
Phylogenetic trees depict the evolutionary relationships among various biological species or entities based on similarities and differences in their physical or genetic characteristics. They imply that taxa joined together in the tree have descended from a common ancestor. Phylogenetic trees are useful in fields like bioinformatics, systematics, and phylogenetic comparative methods. However, phylogenetic trees have limitations as they do not necessarily accurately represent evolutionary history and can be confounded by factors like genetic recombination and horizontal gene transfer.