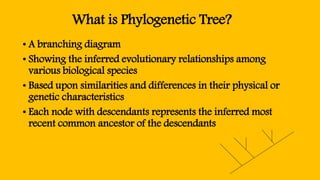
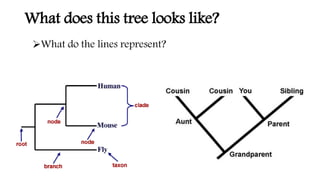
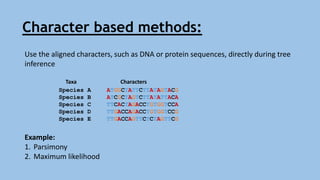
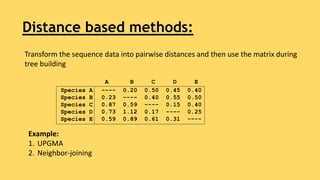
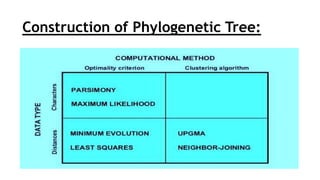
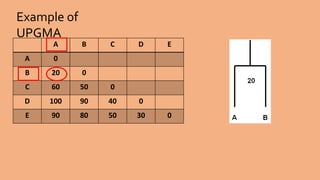
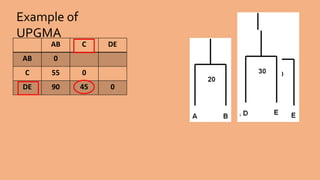
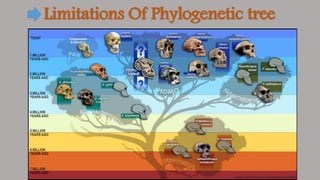
Phylogenetic trees are branching diagrams that show the evolutionary relationships among various biological species based on similarities and differences in their physical or genetic characteristics. Each node represents the inferred most recent common ancestor of the descendants. Phylogenetic trees are constructed using character-based or distance-based methods and can be used to understand human and animal origins, molecular evolution processes, and the spread of disease.