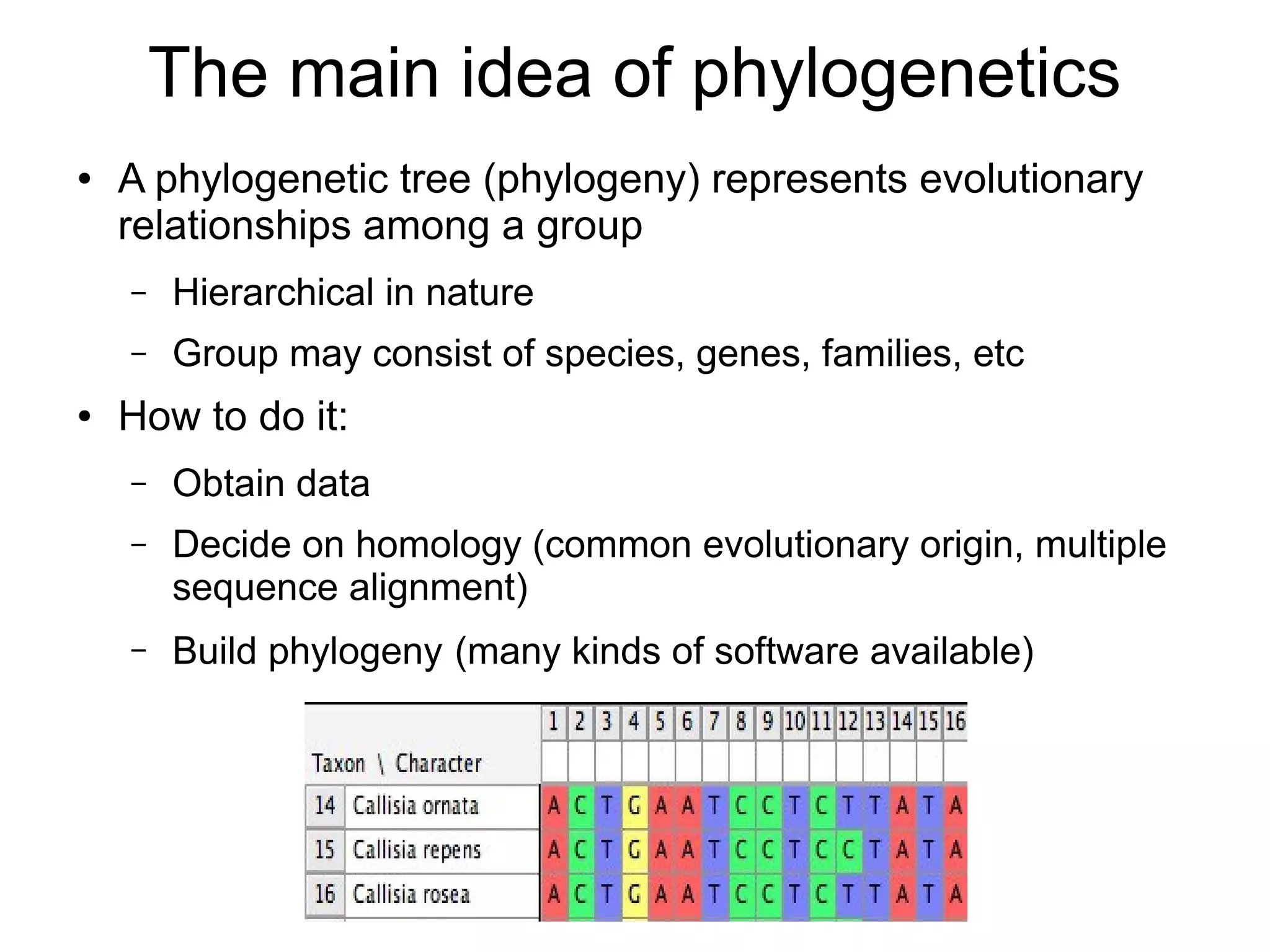
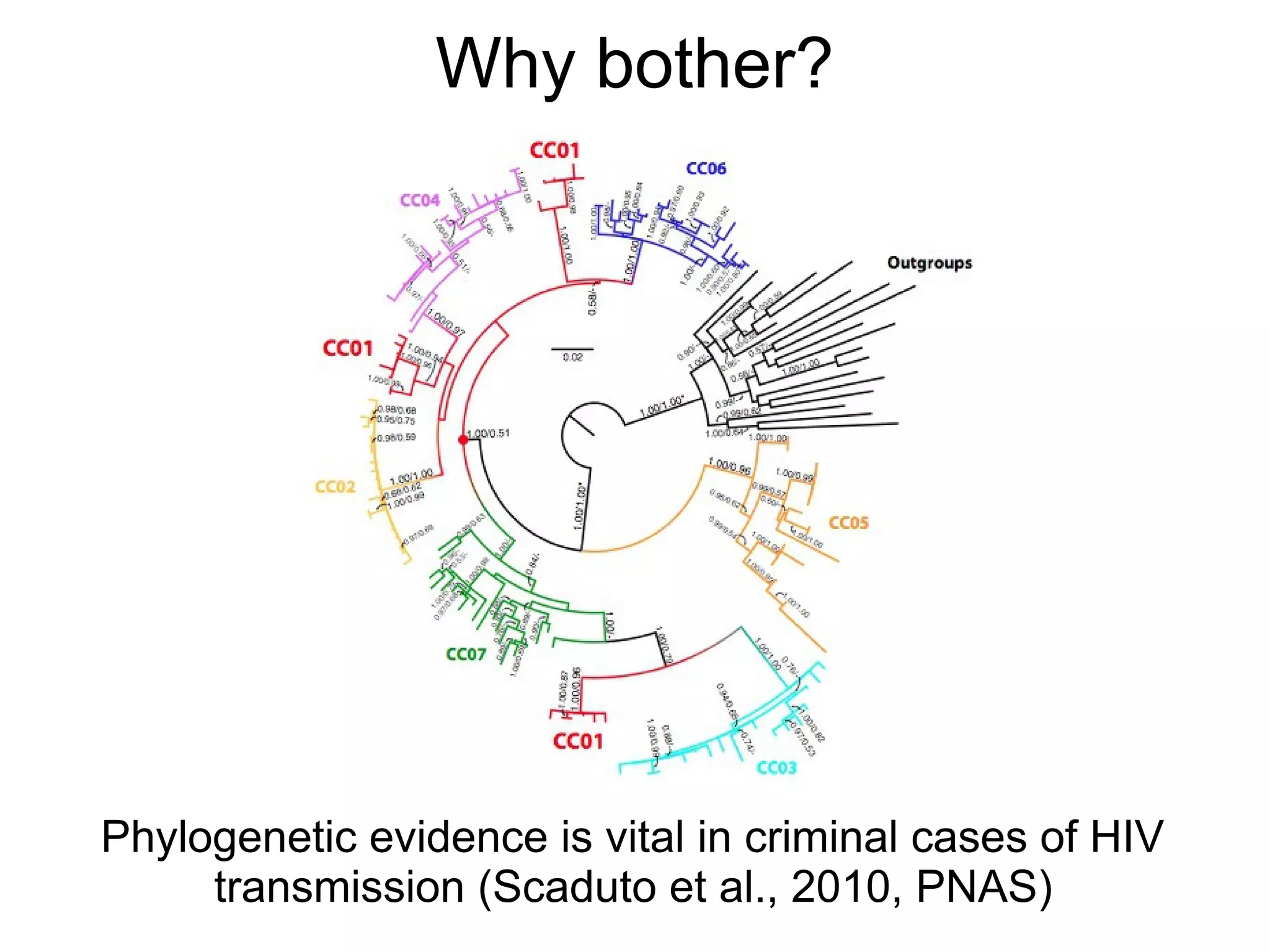
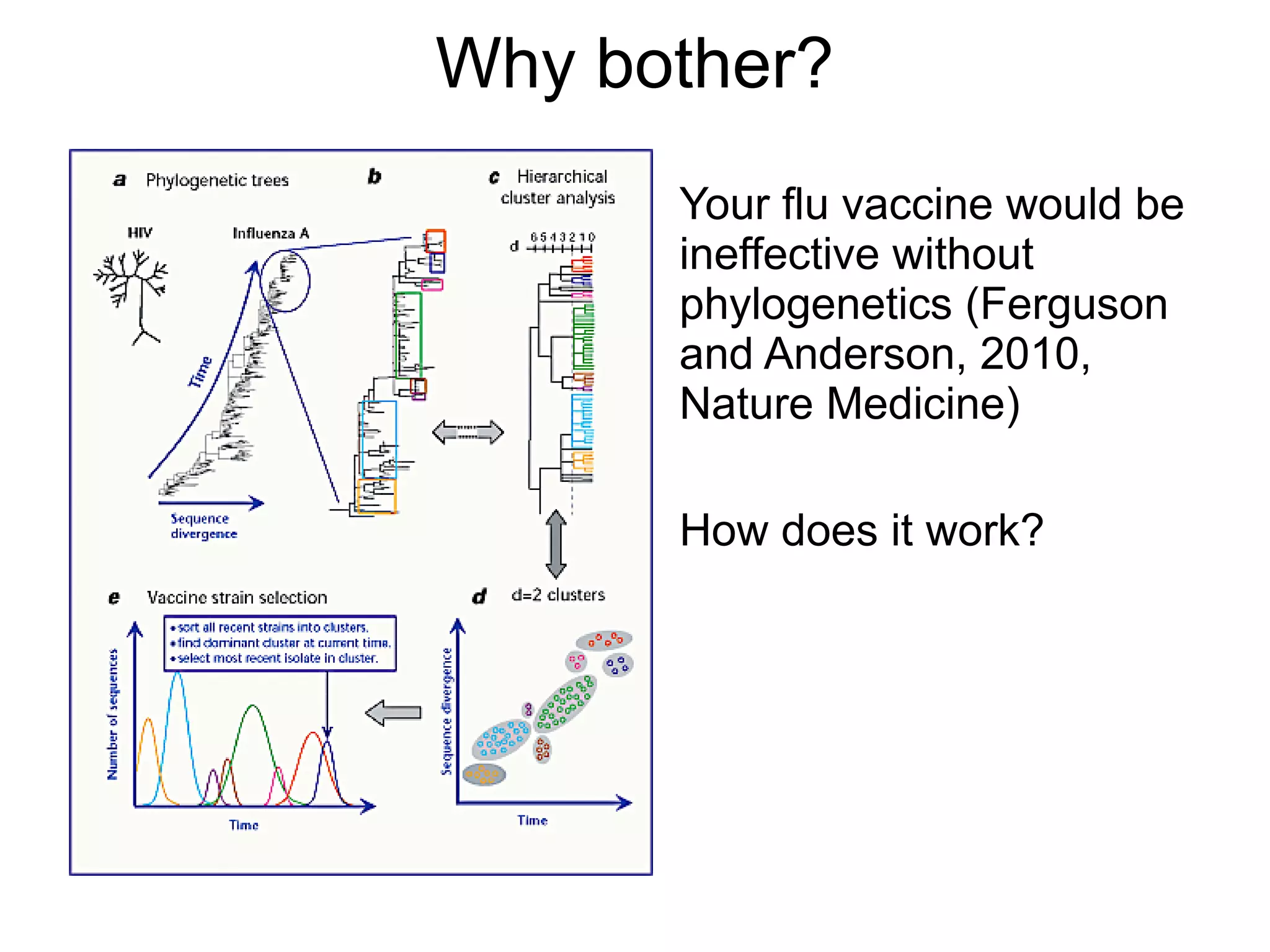
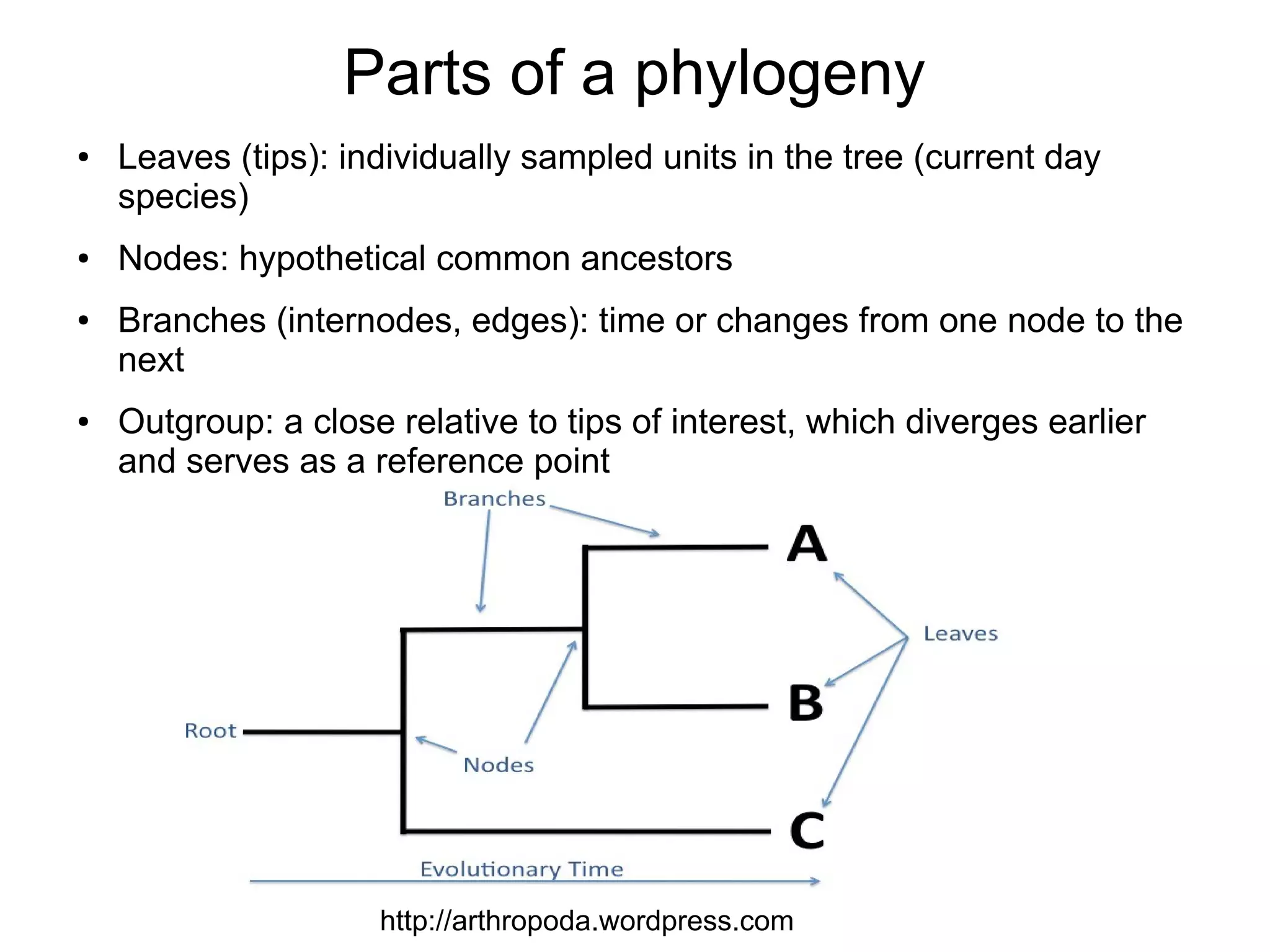
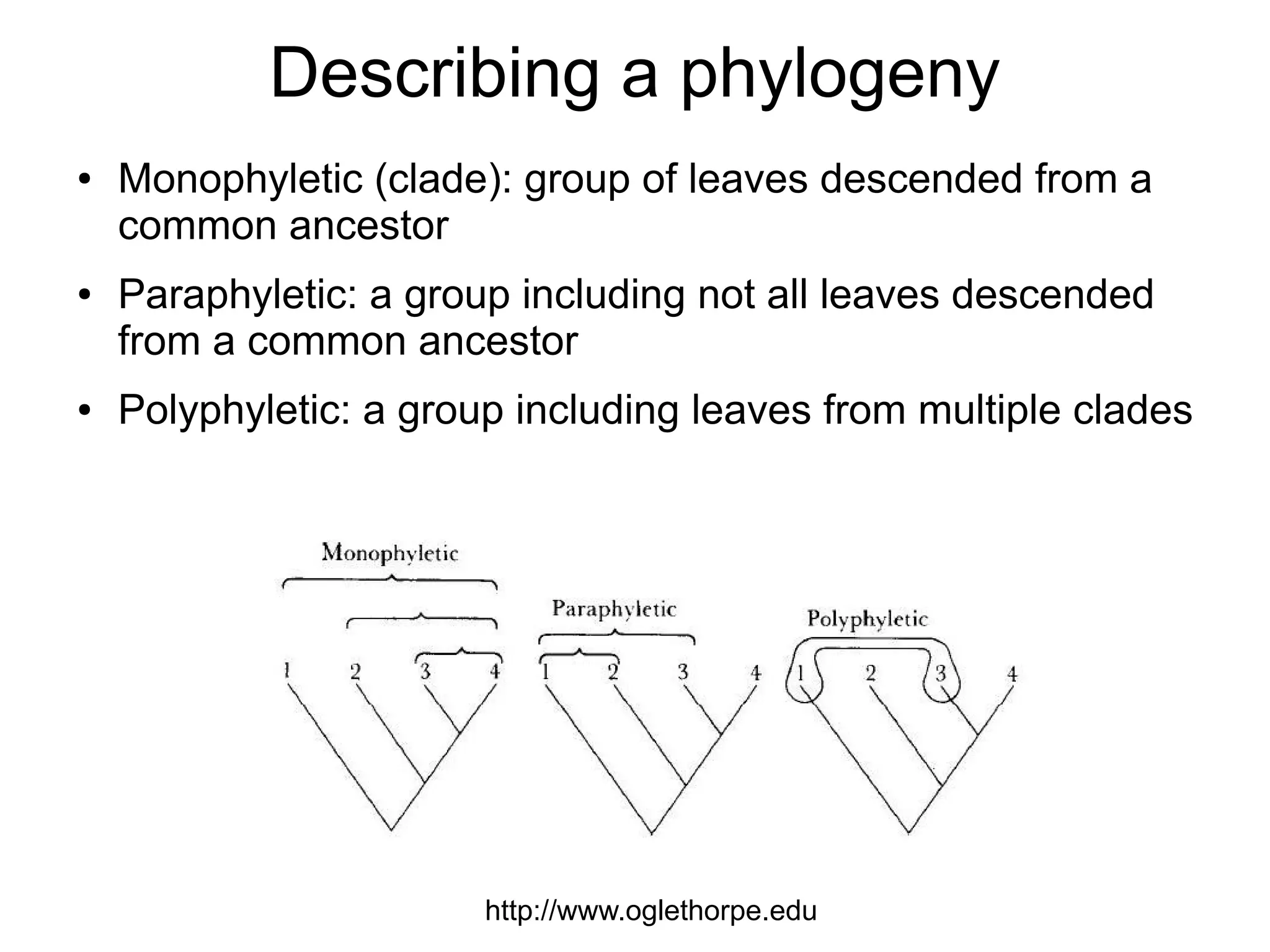
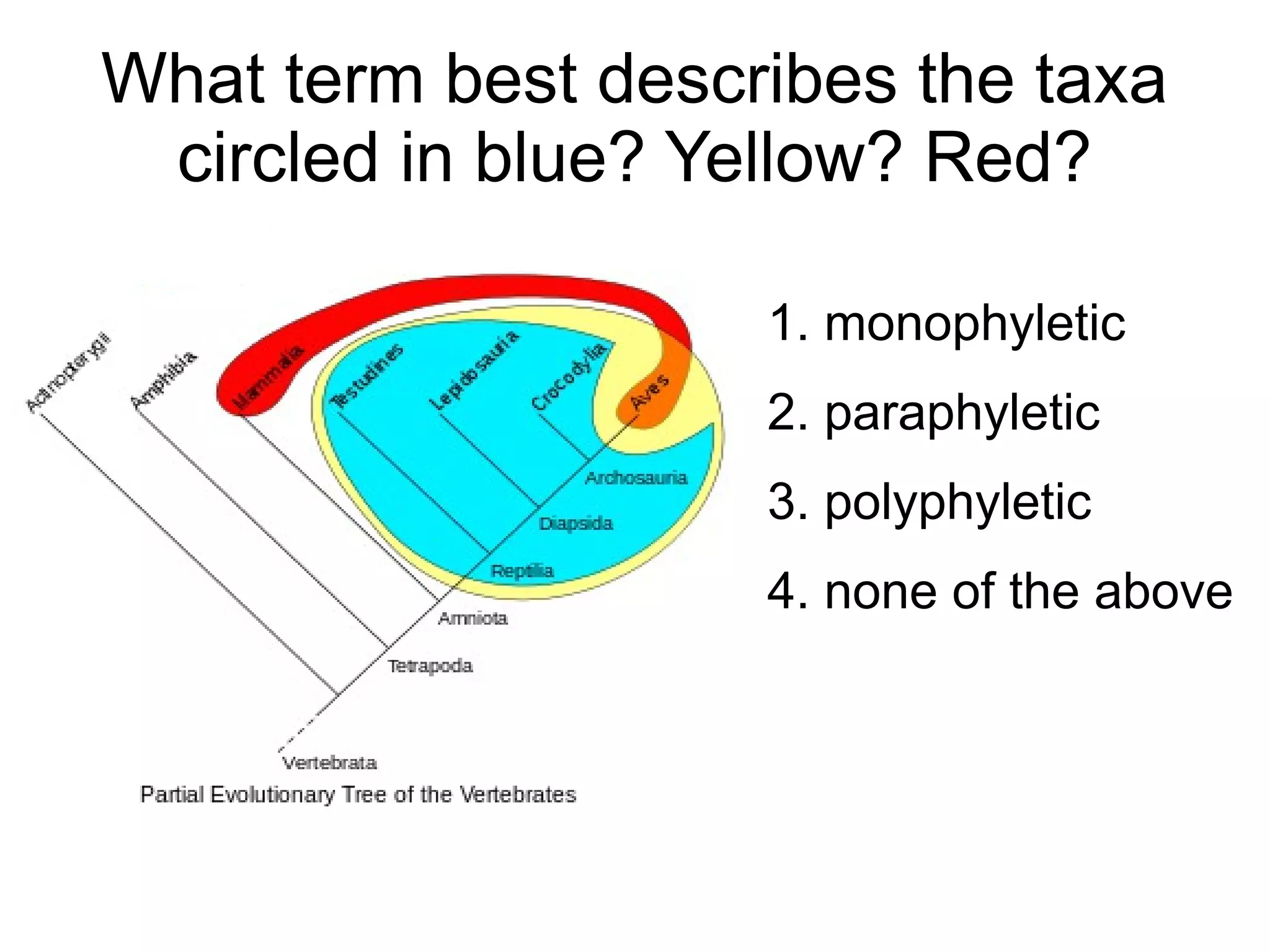
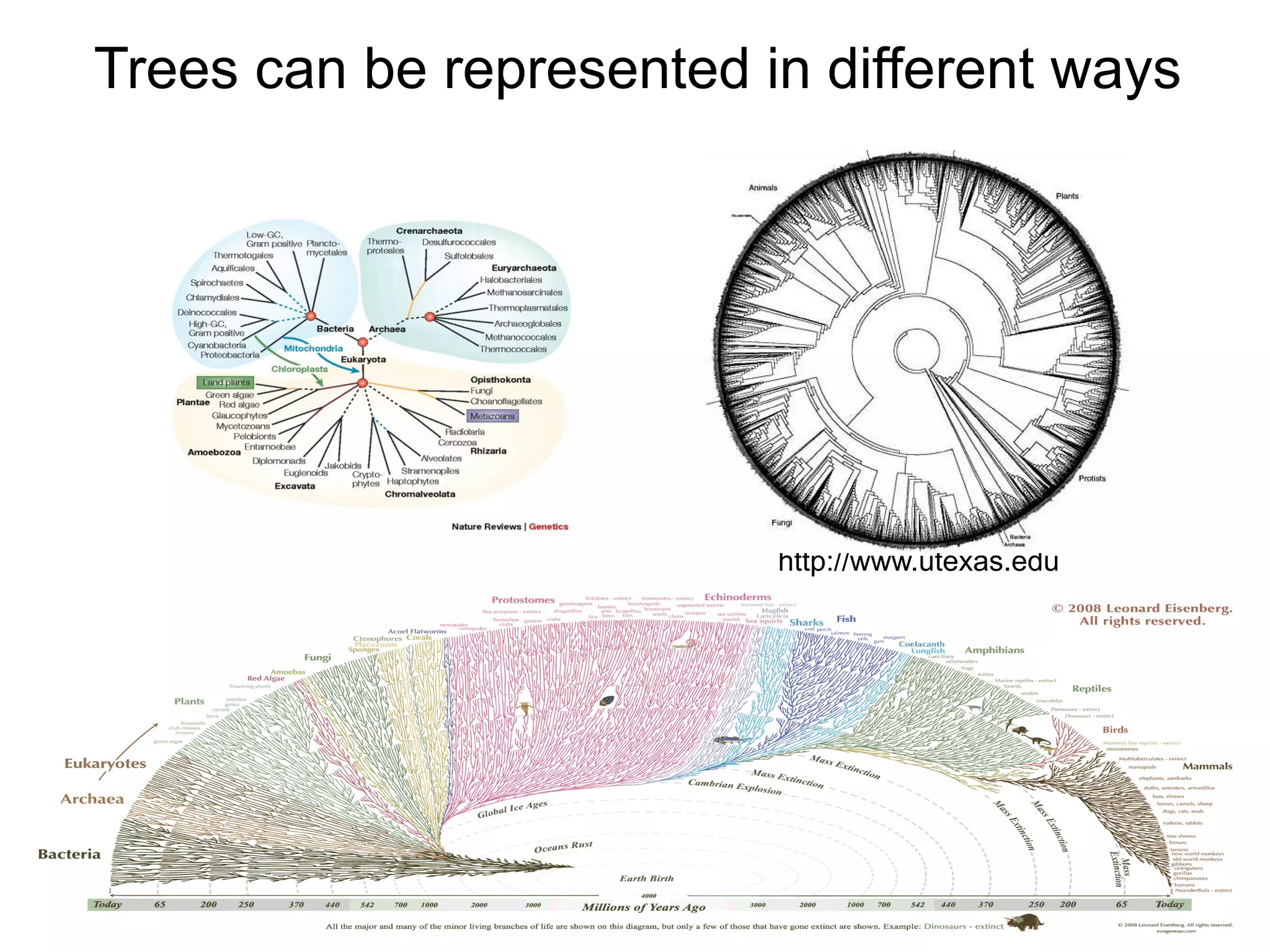
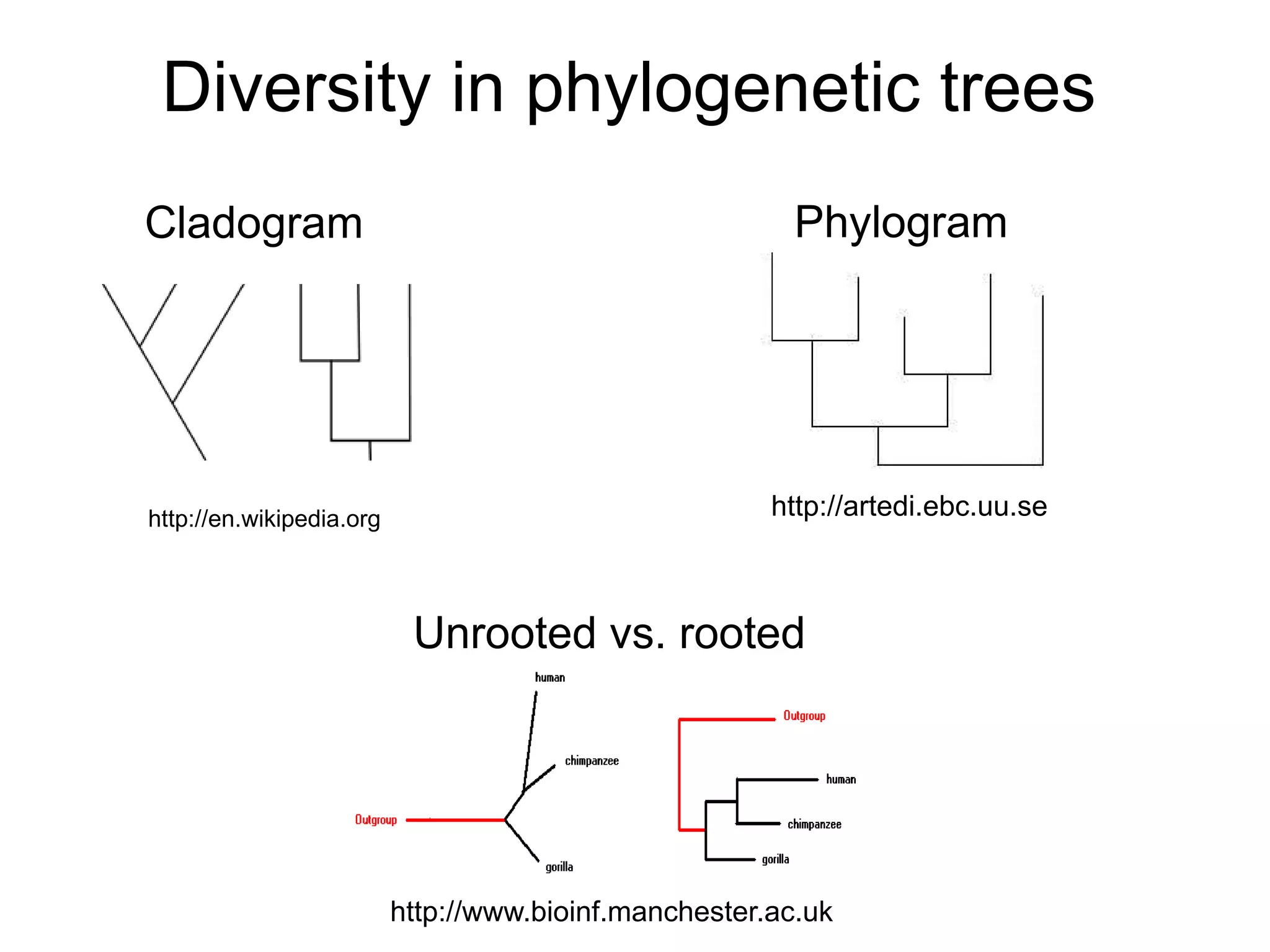
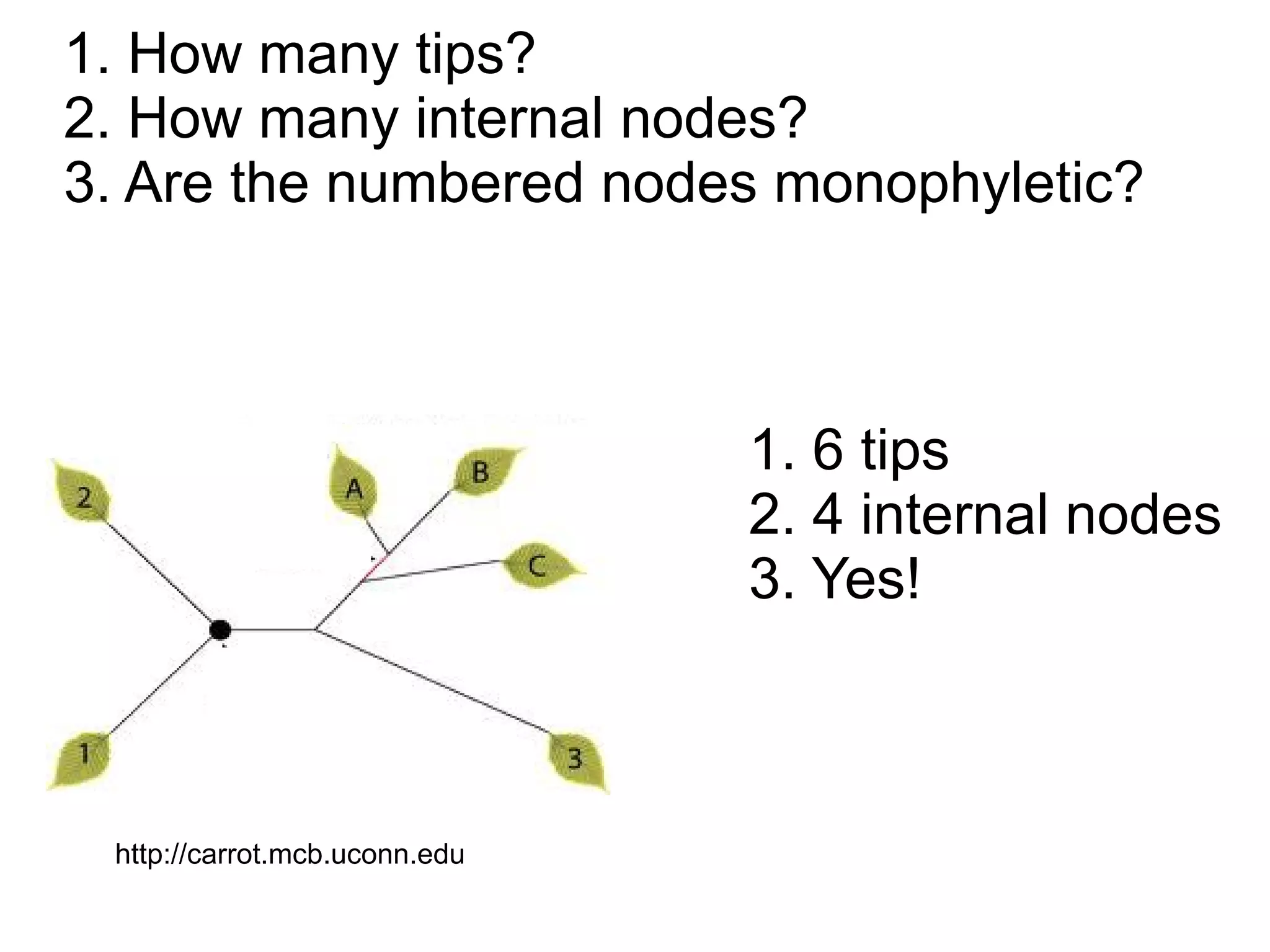
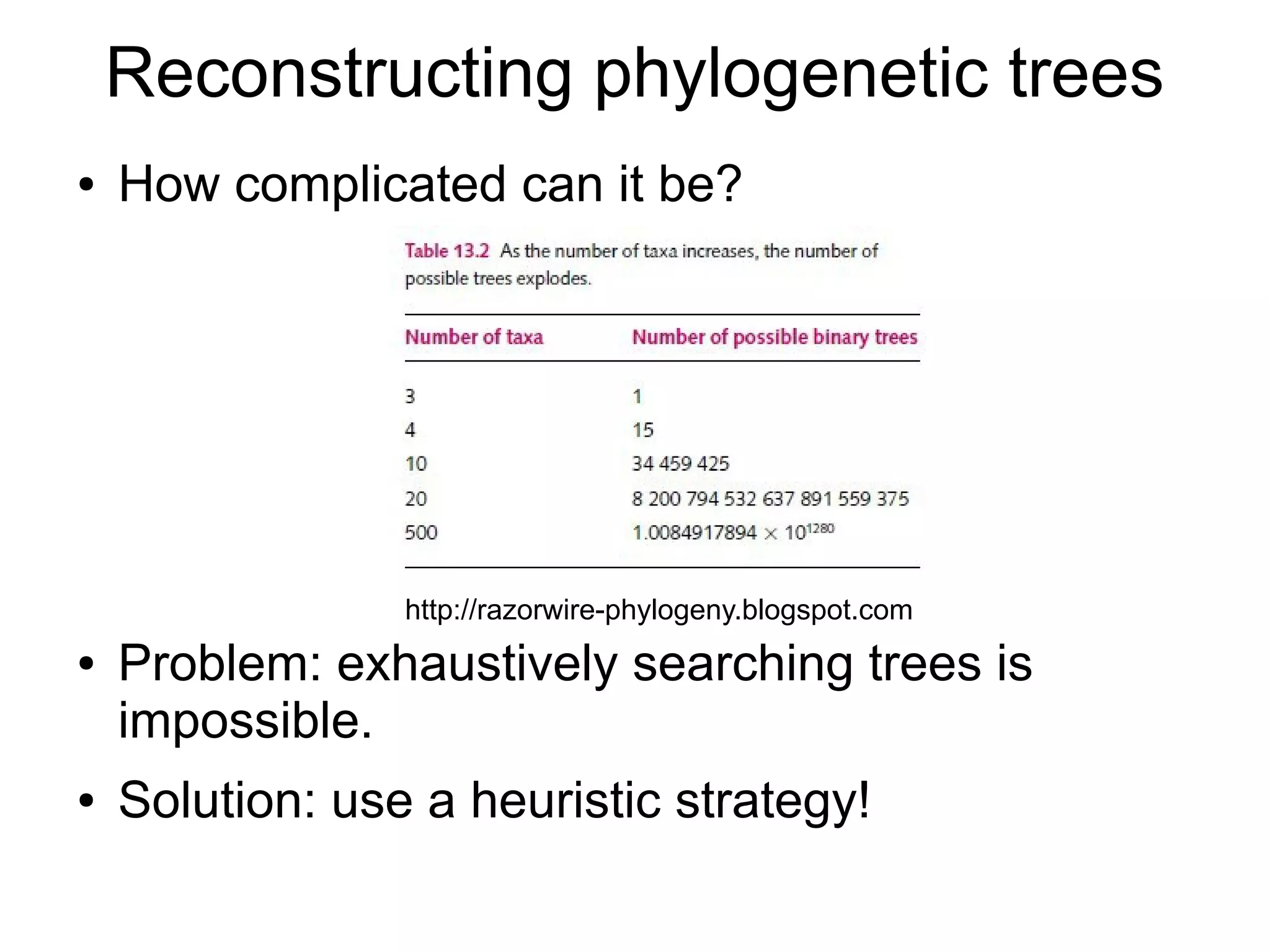
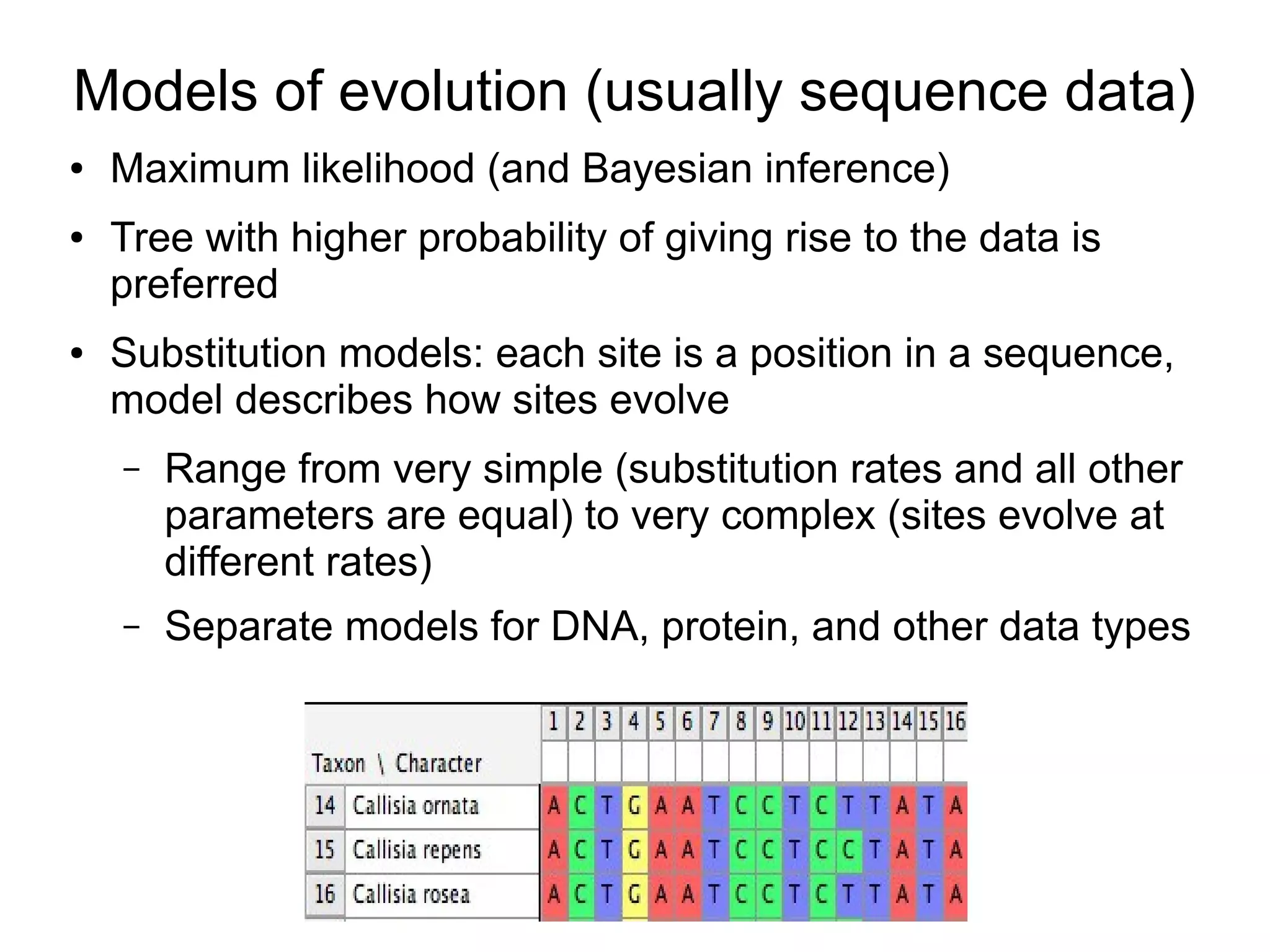
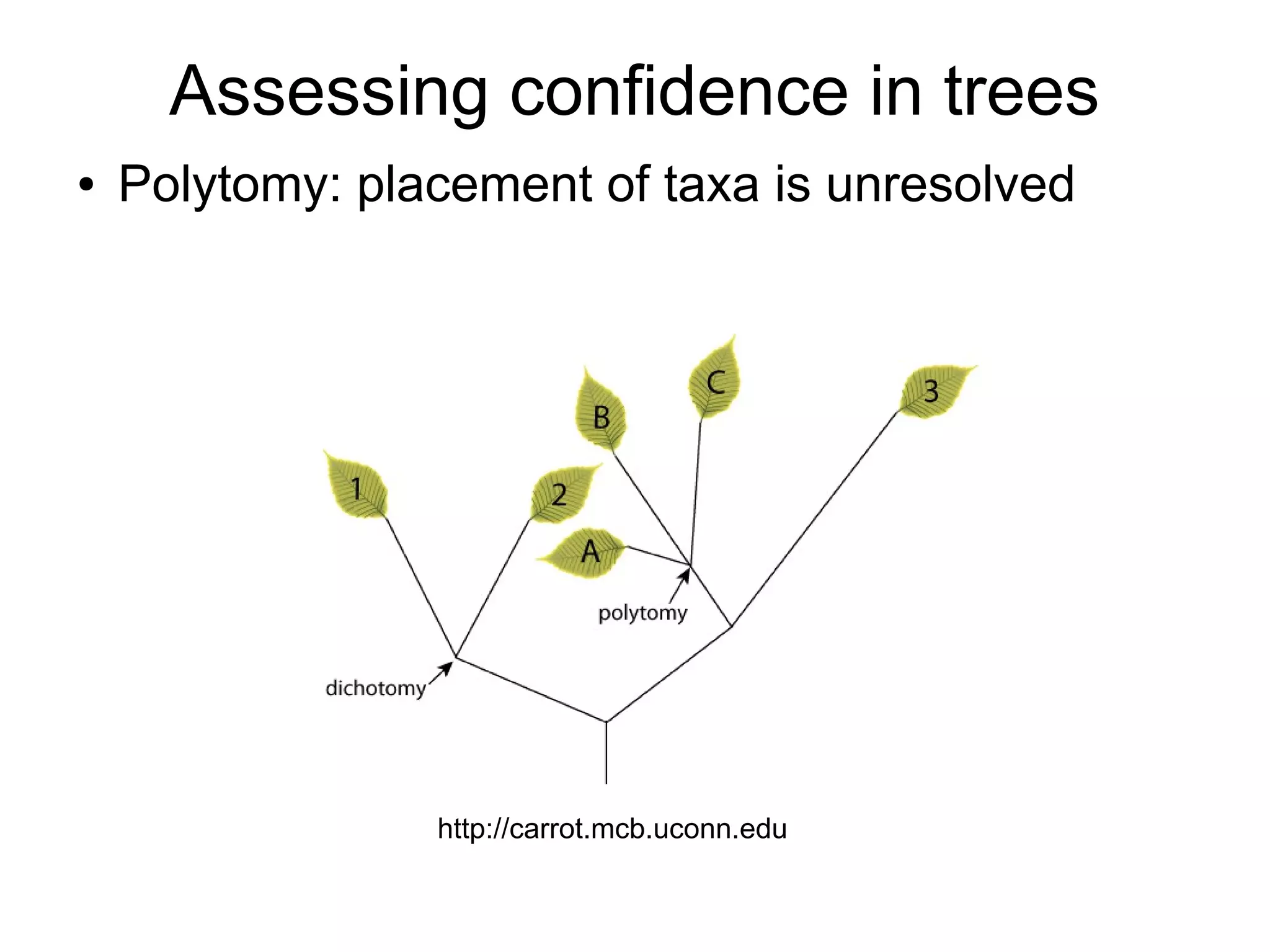
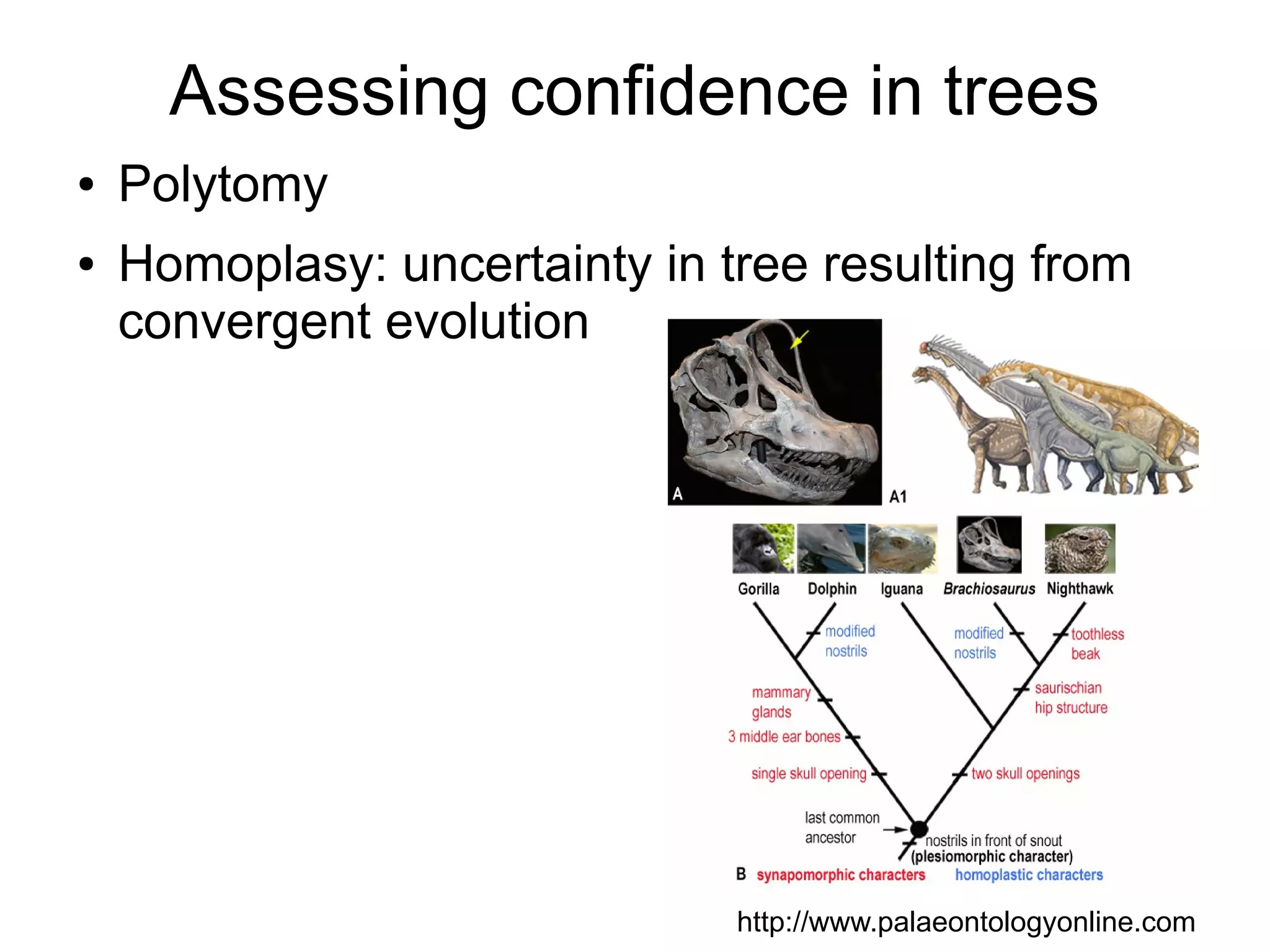
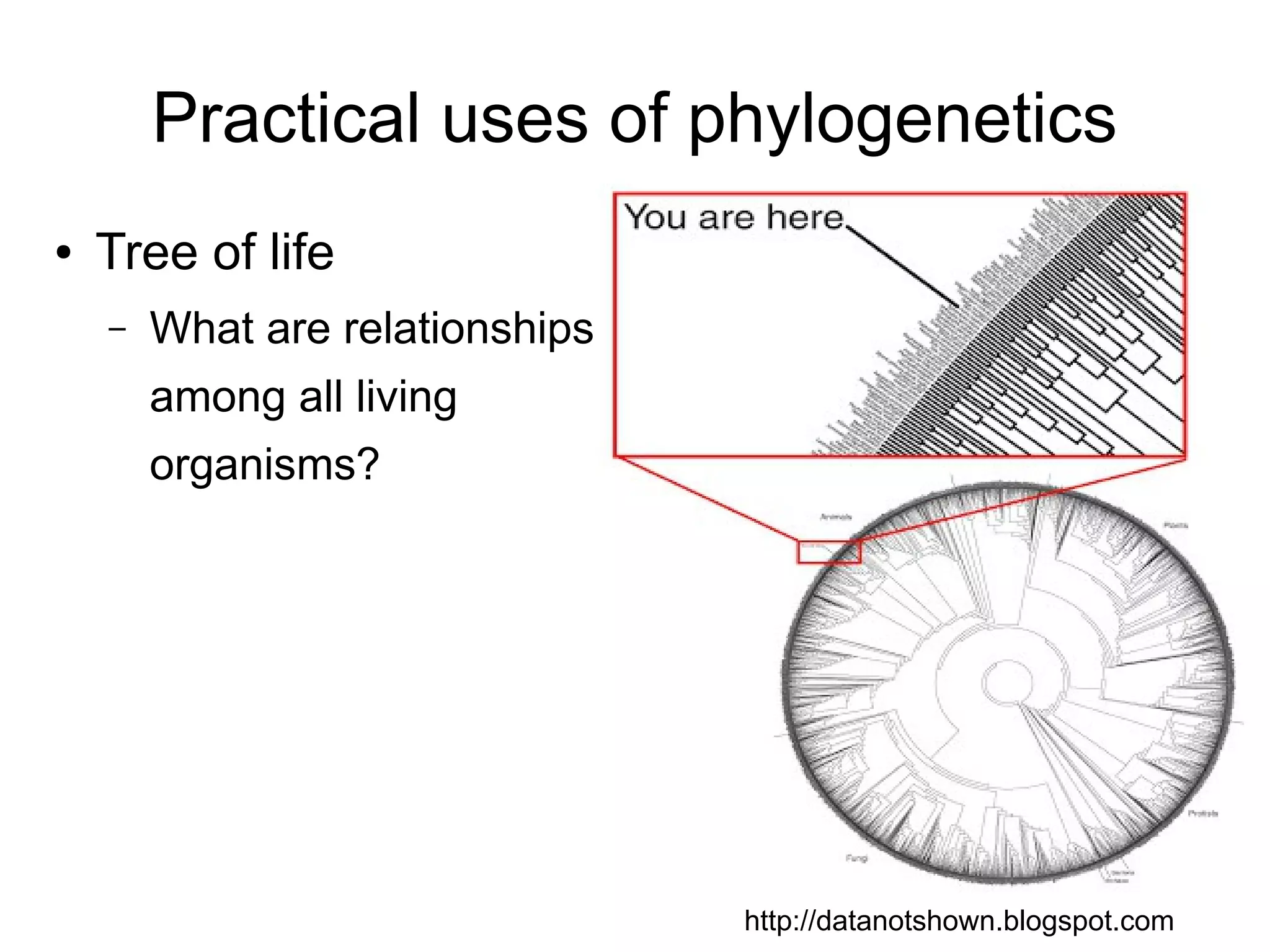
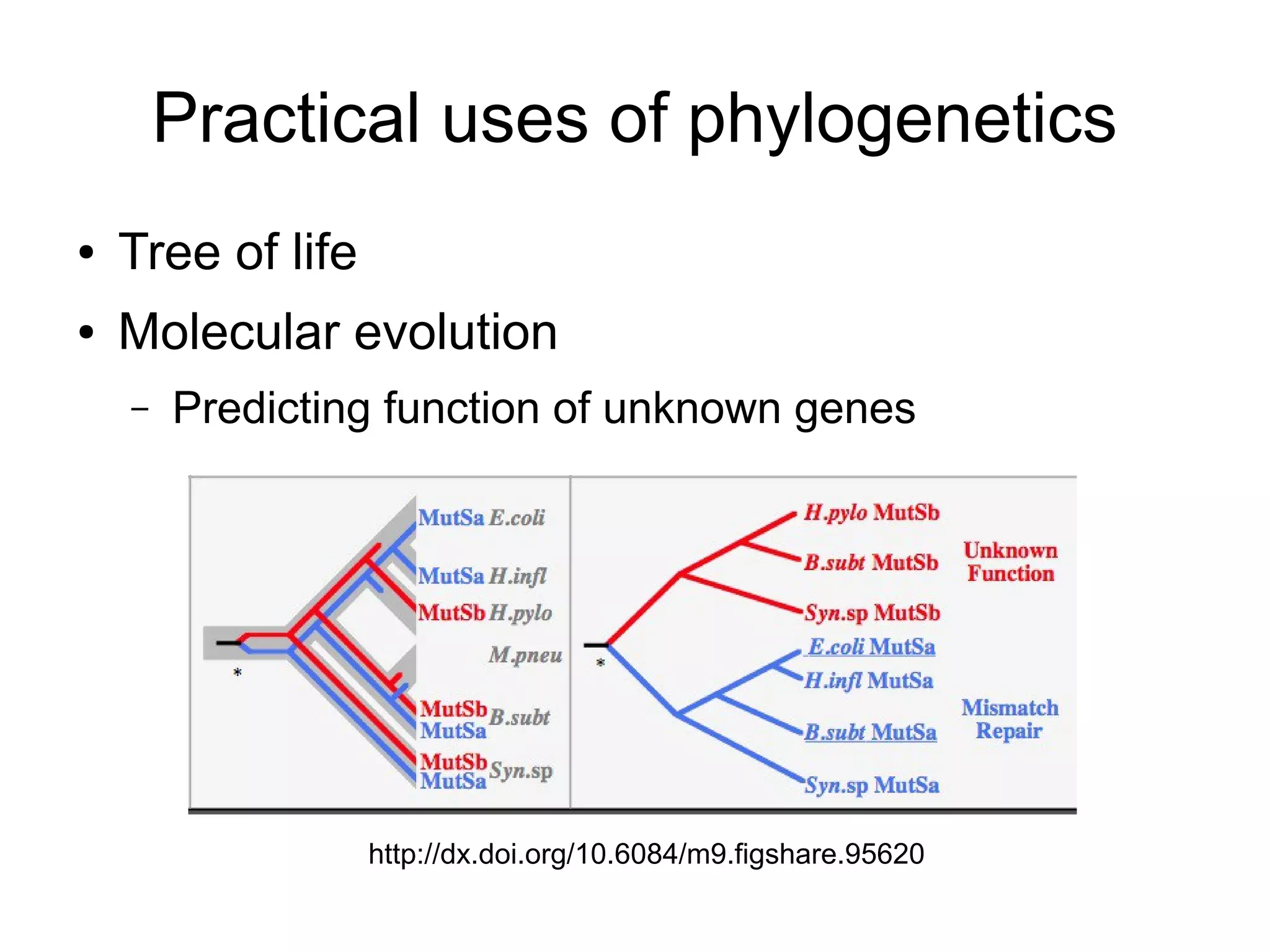
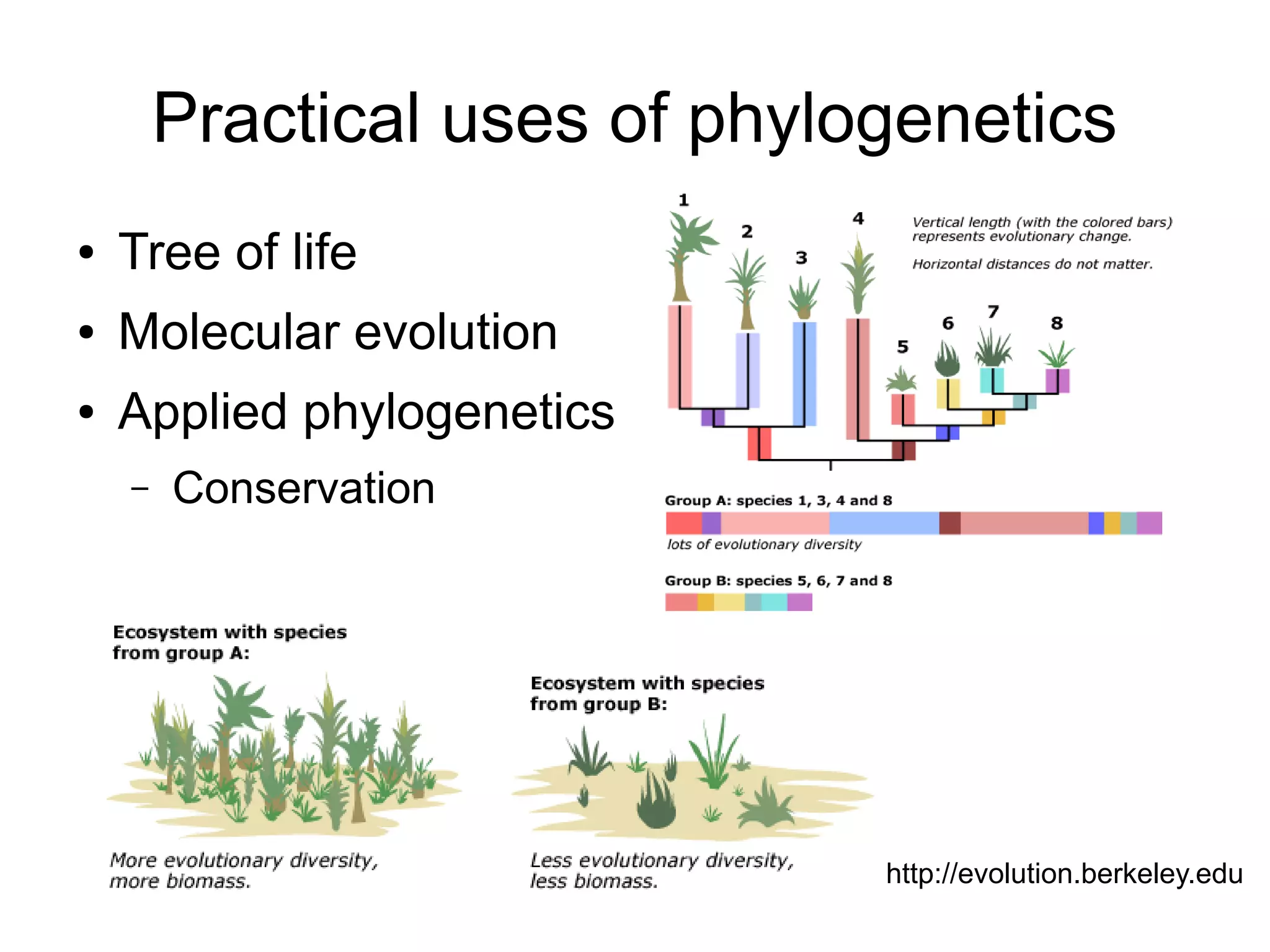
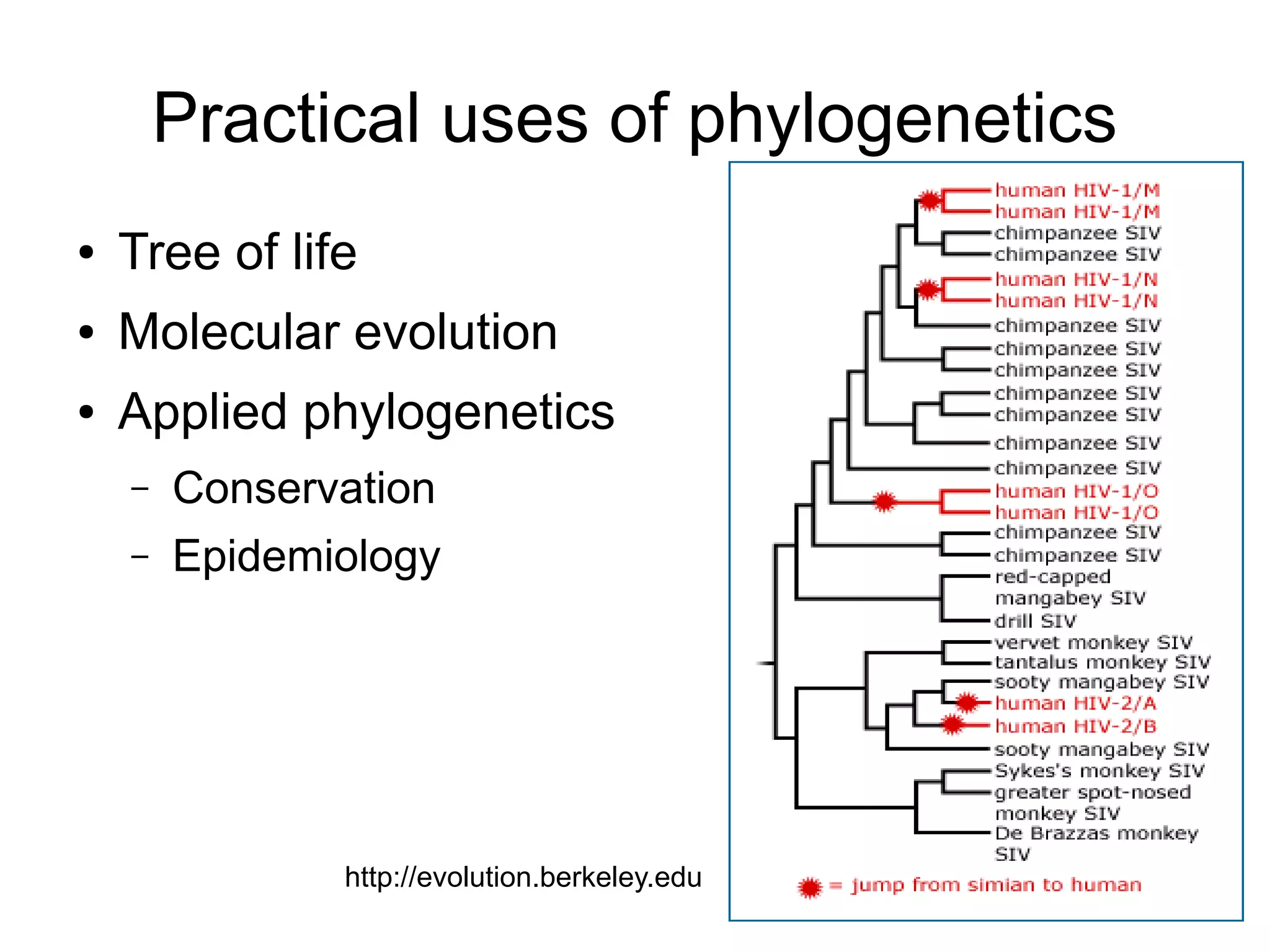
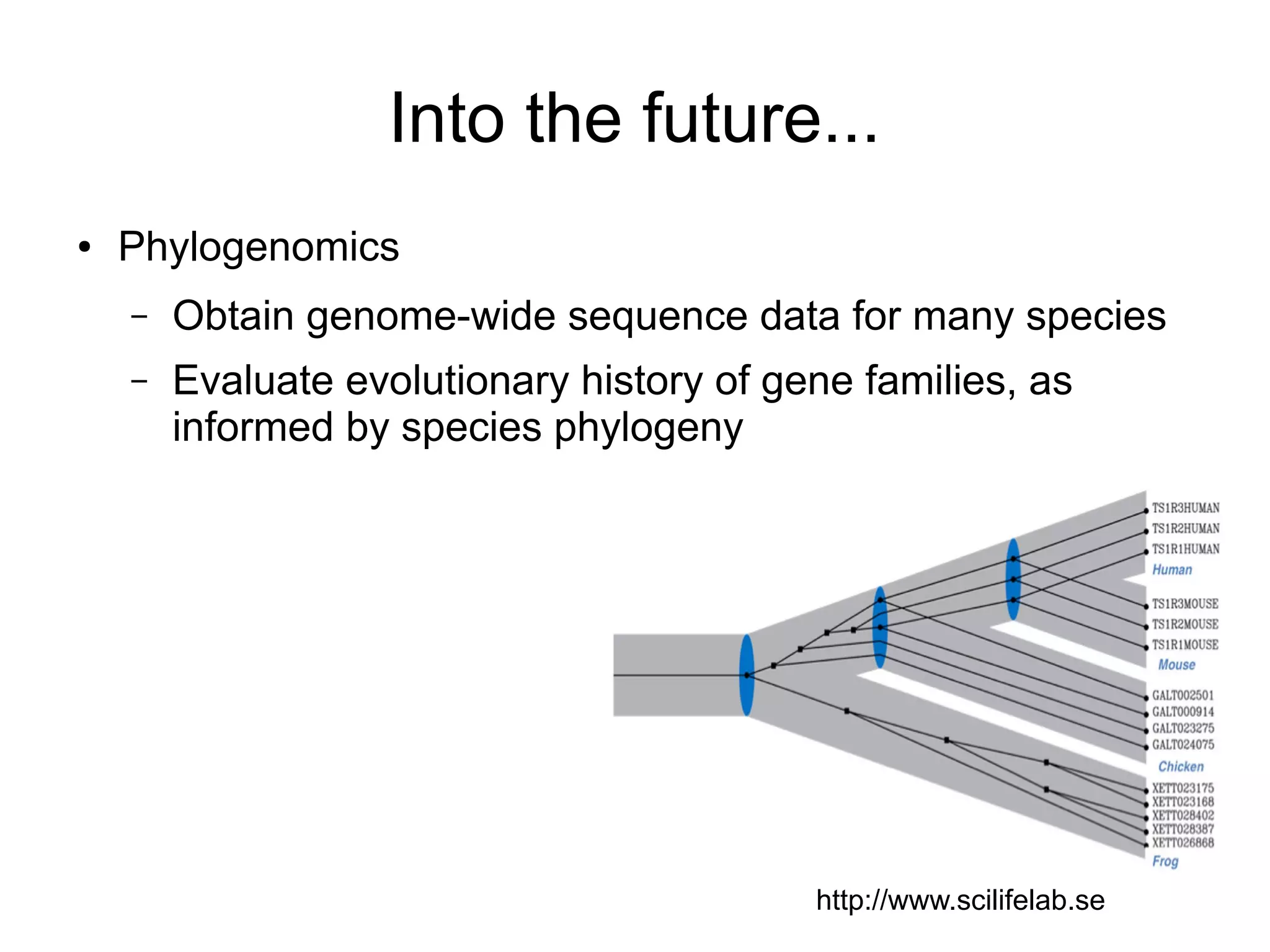
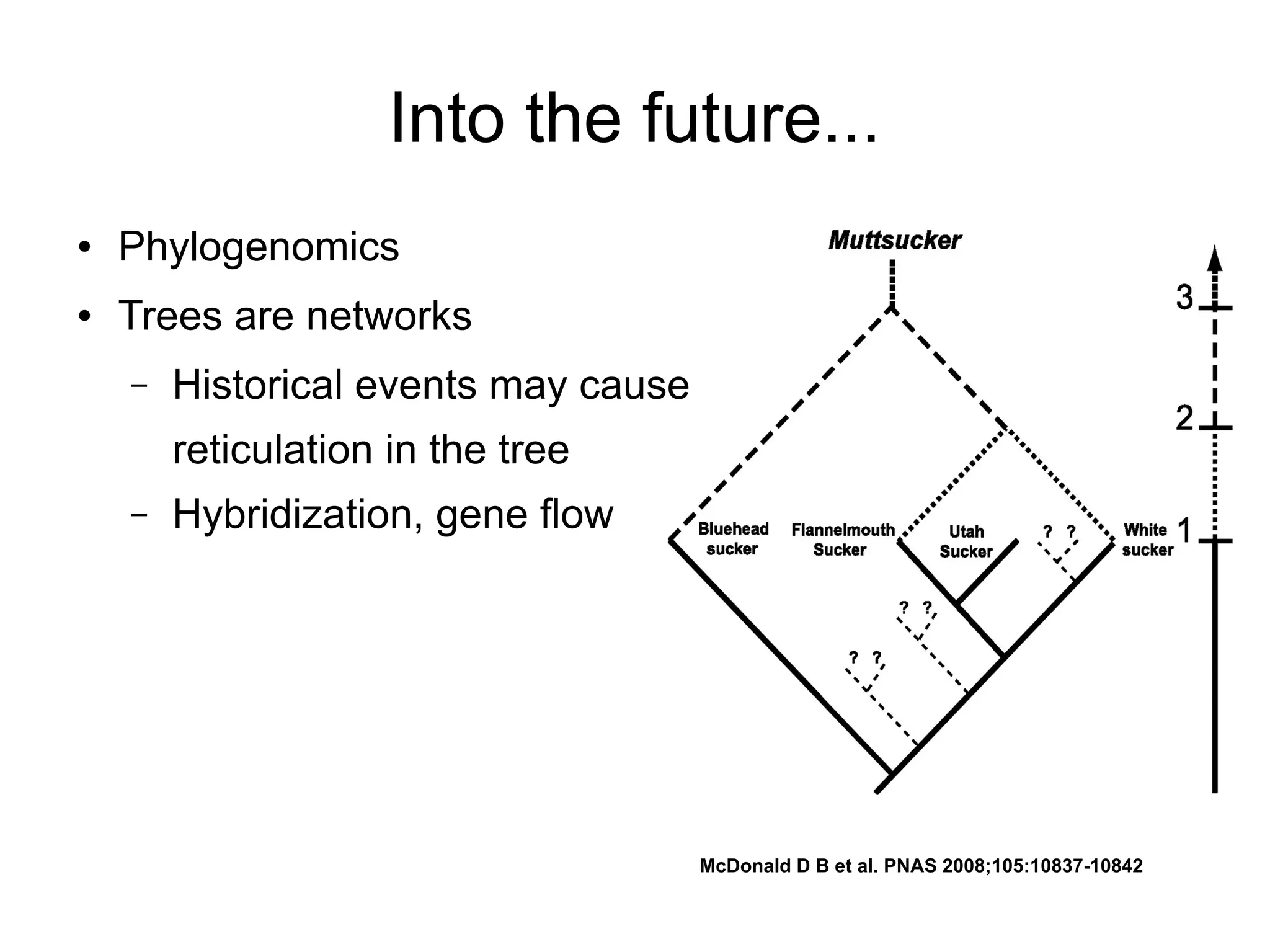
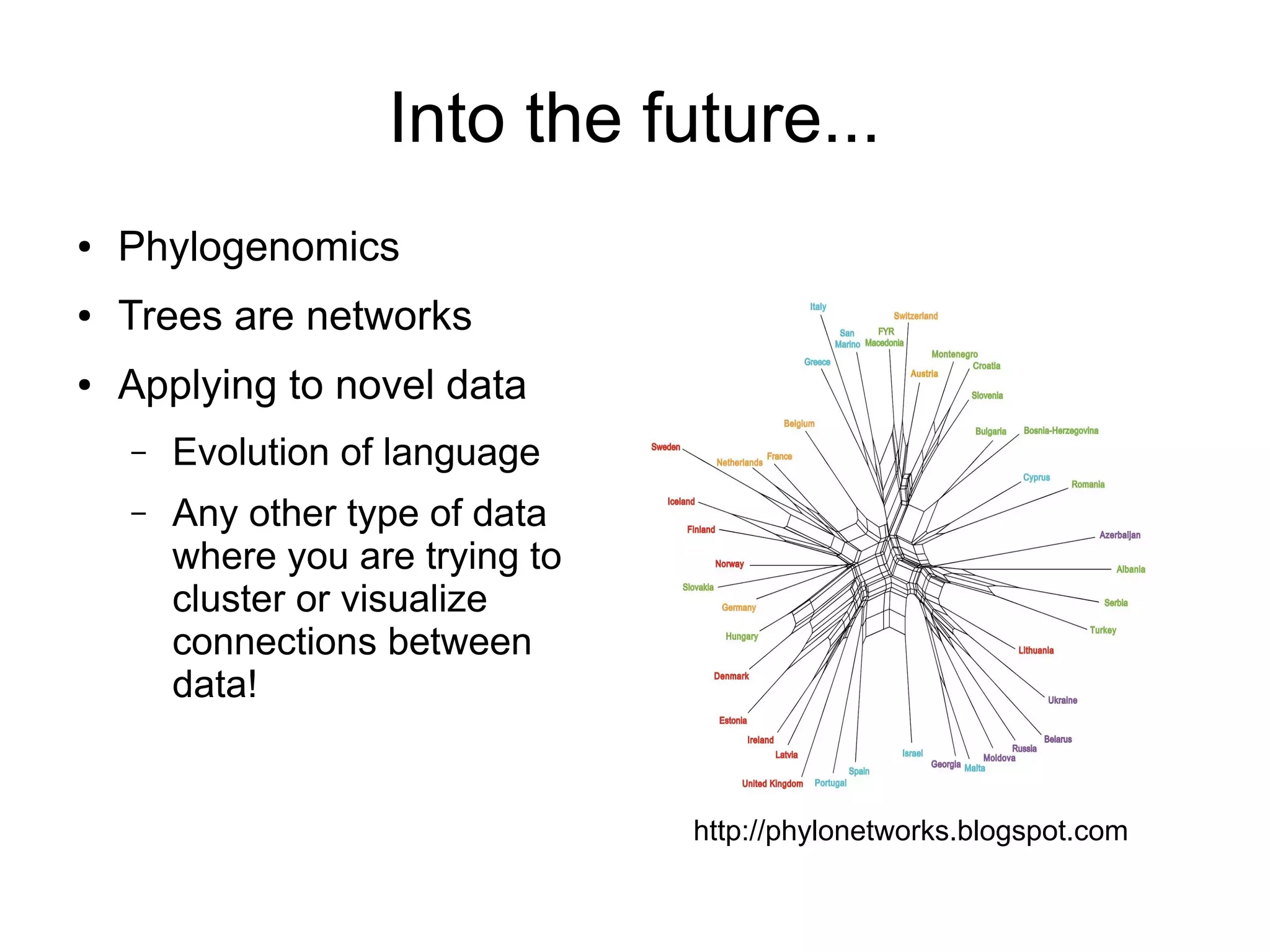
Phylogenetics is the study of evolutionary relationships among groups of organisms. A phylogenetic tree represents these relationships, showing how groups are hierarchically related through hypothetical common ancestors. Building phylogenies involves obtaining data, determining homology, and using software to construct the tree based on optimality criteria like parsimony or likelihood. Phylogenies have many practical uses like understanding the tree of life, molecular evolution, conservation, and epidemiology. Future directions include phylogenomics using genome-wide data and considering trees as networks rather than hierarchies.