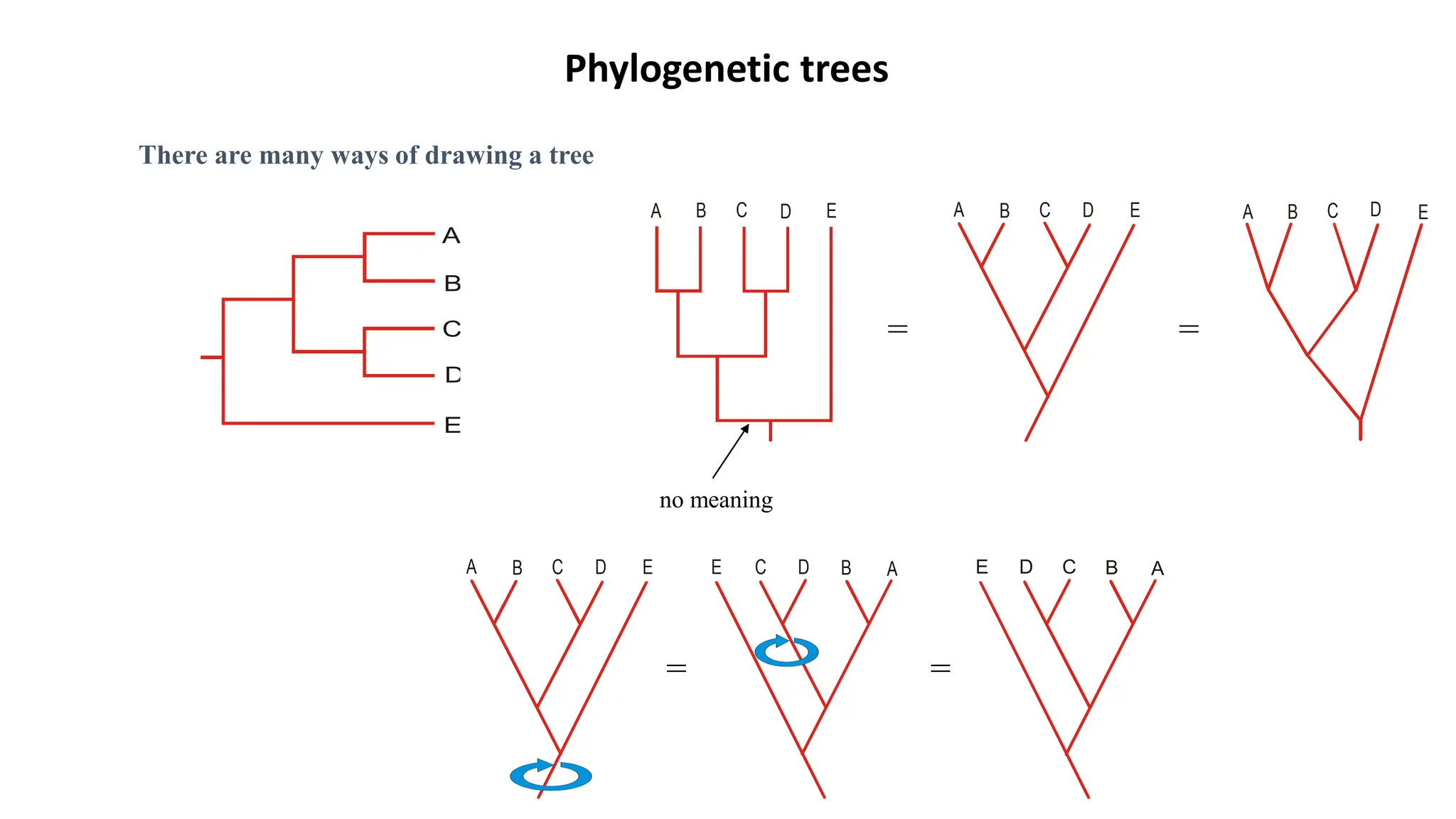
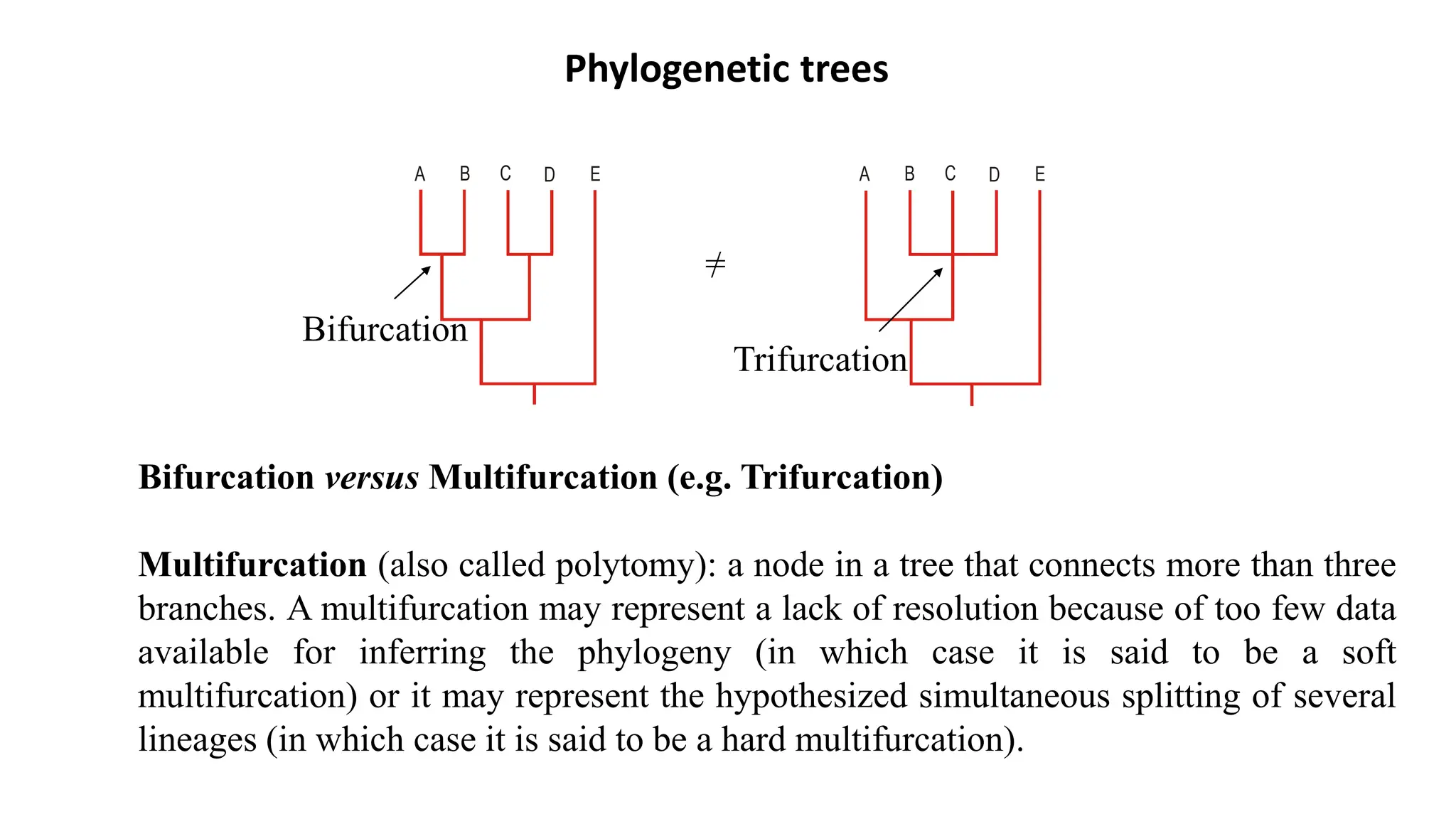
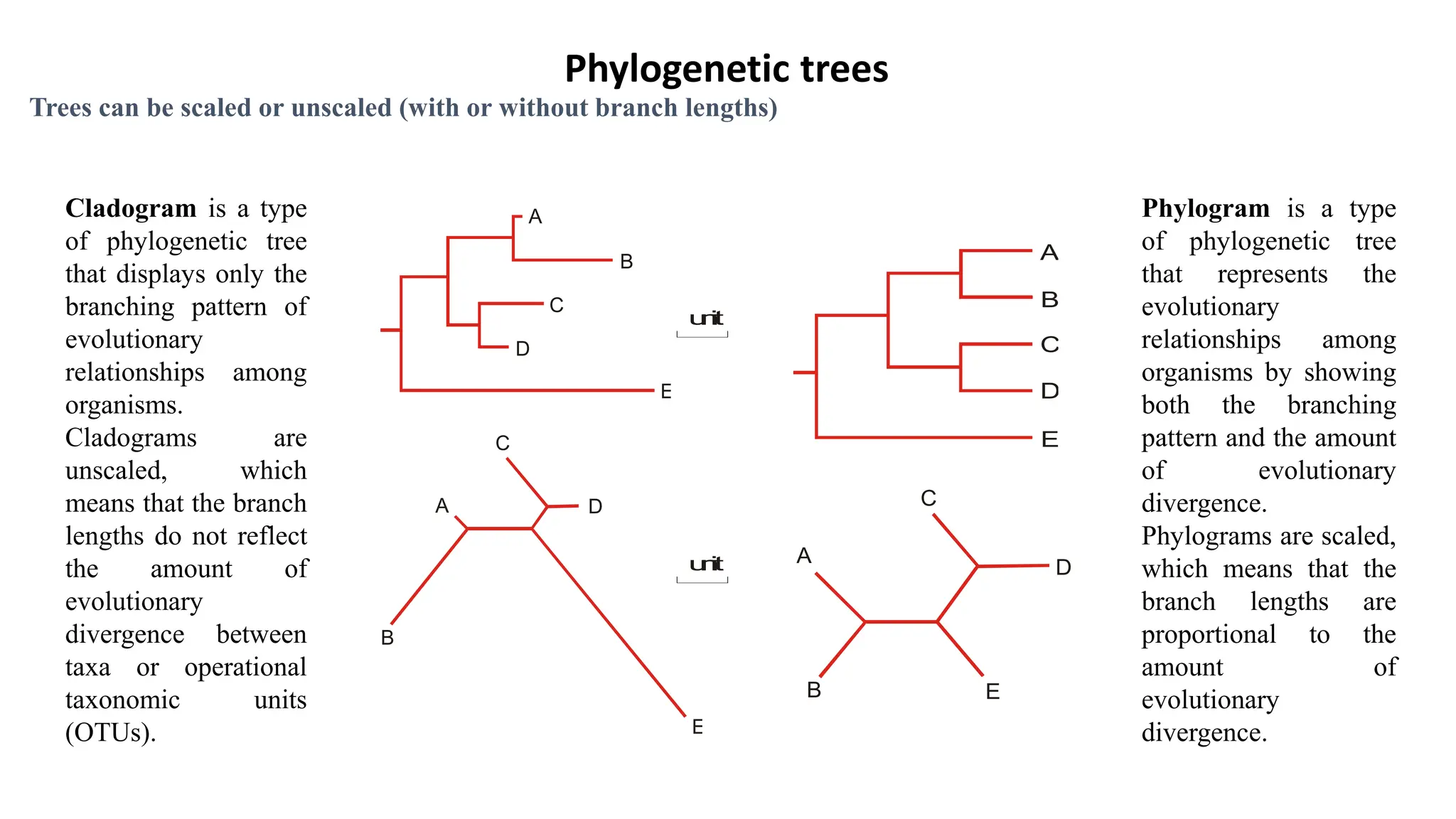
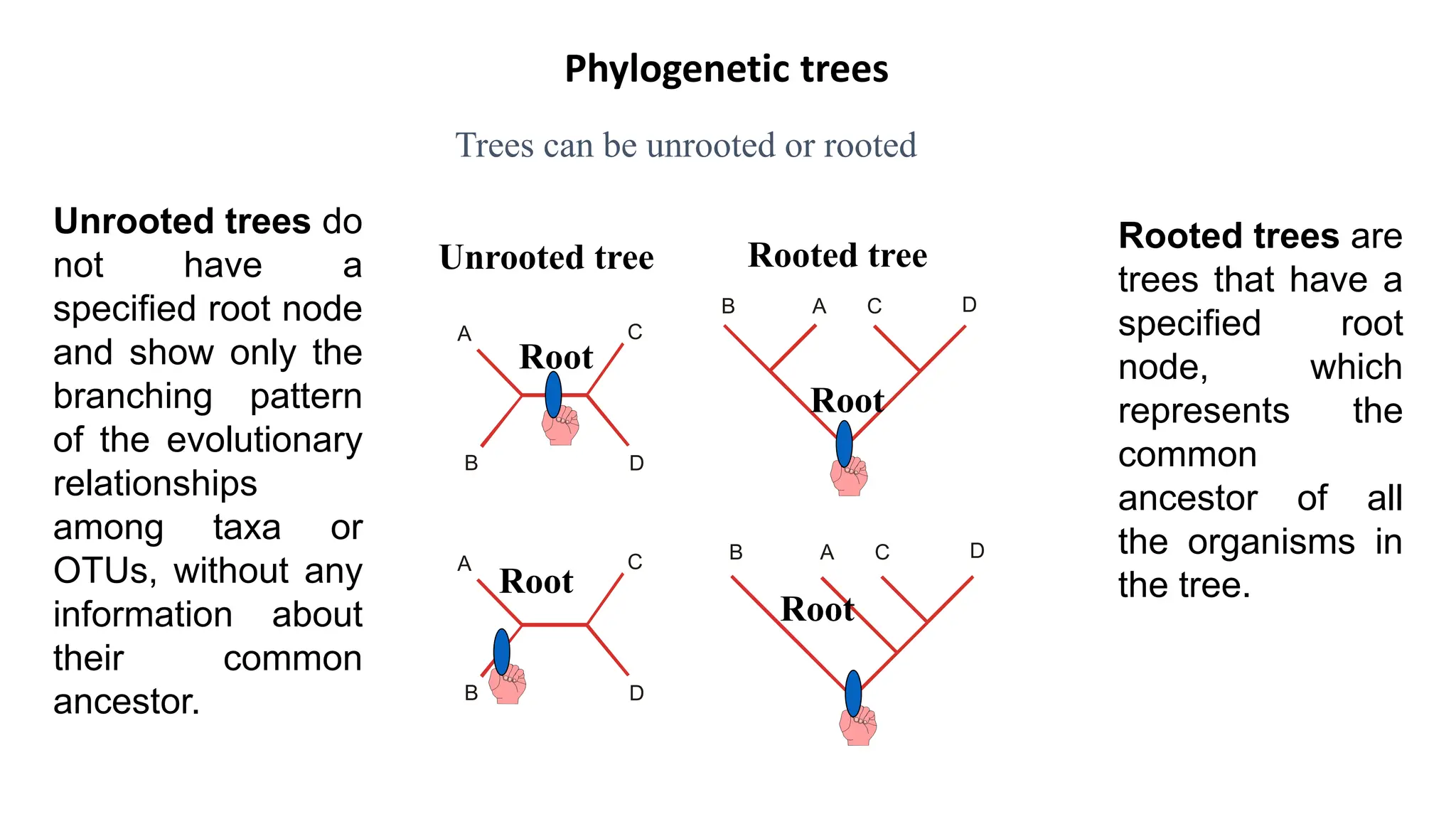
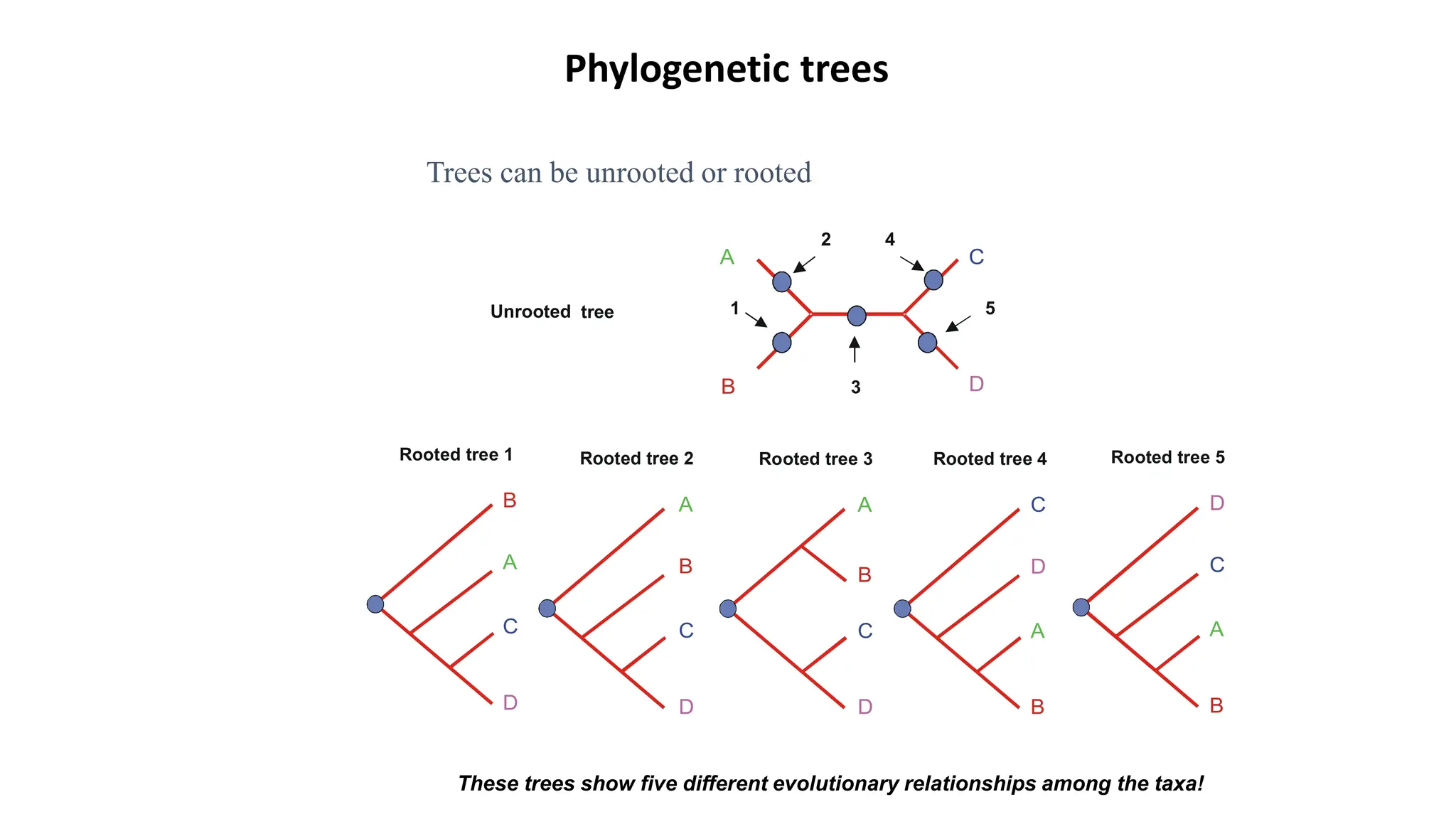
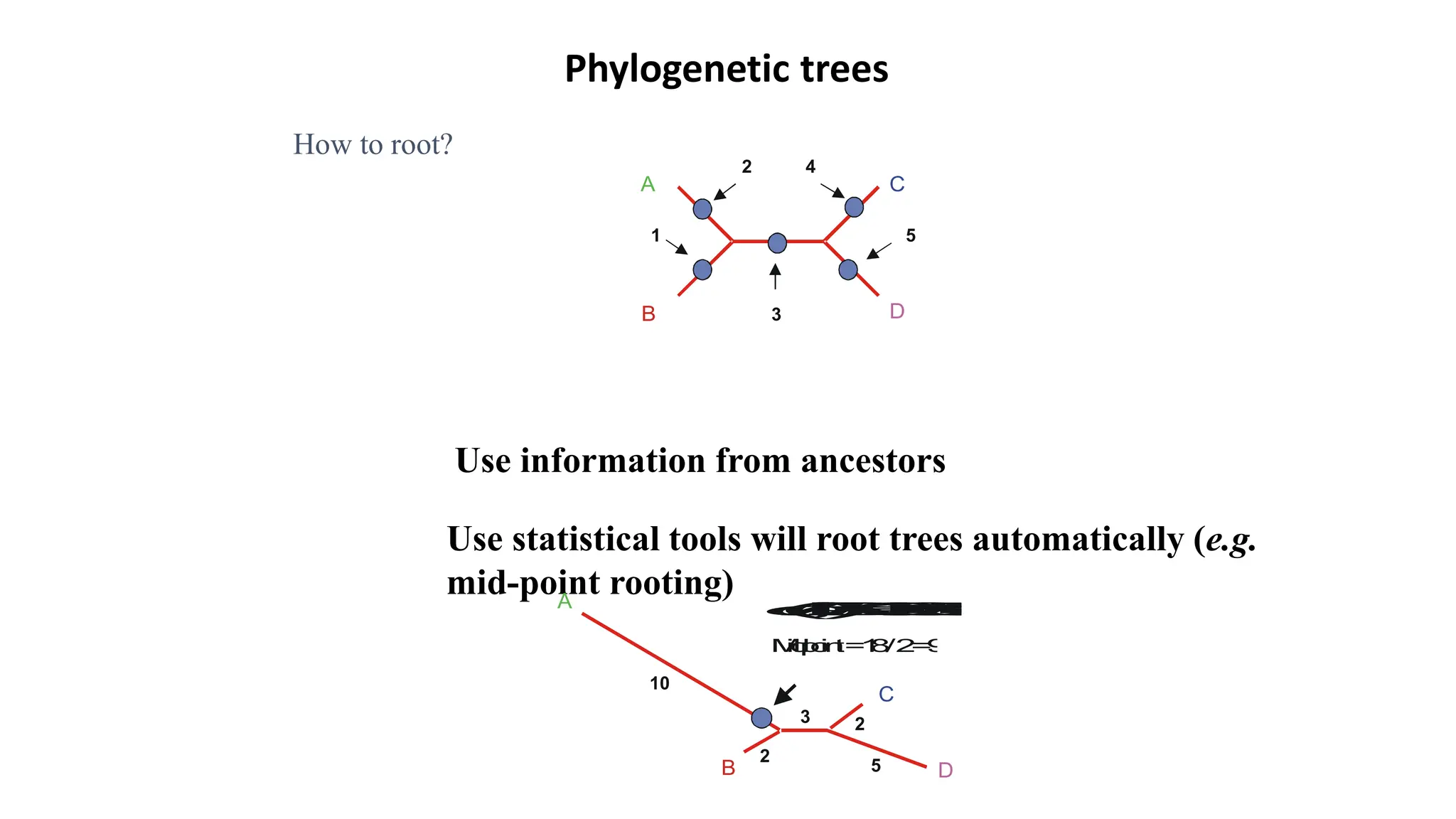
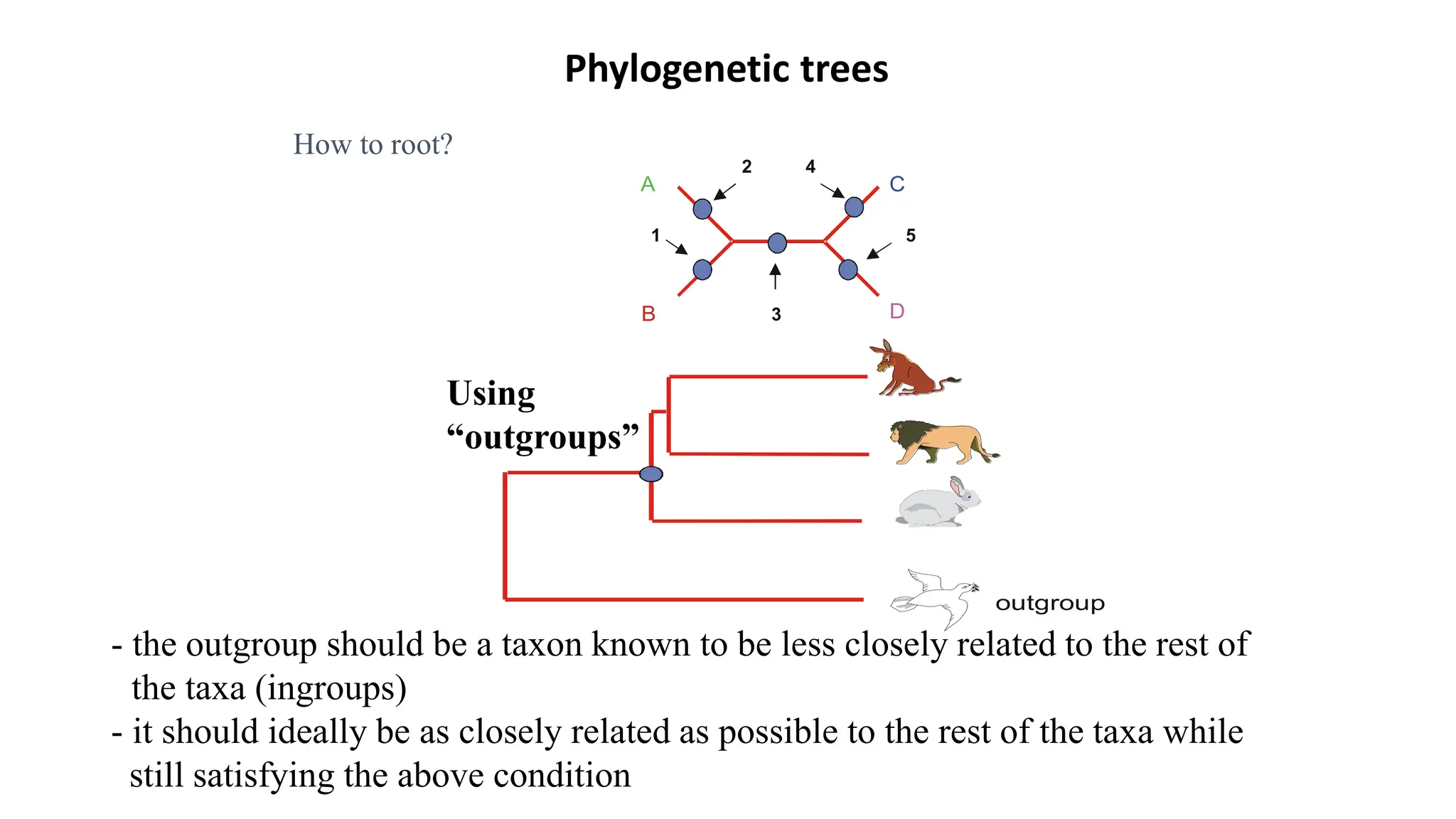
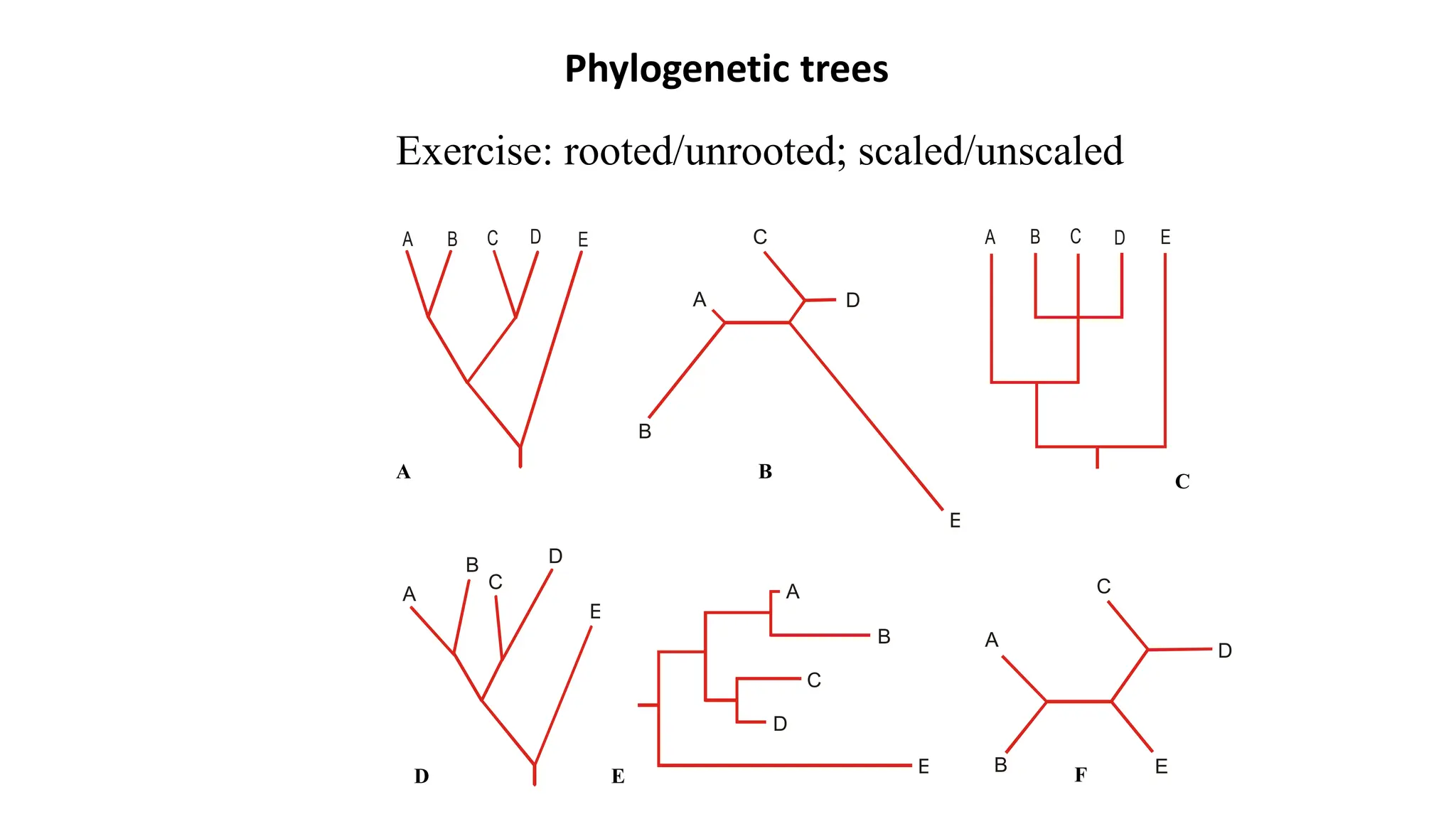
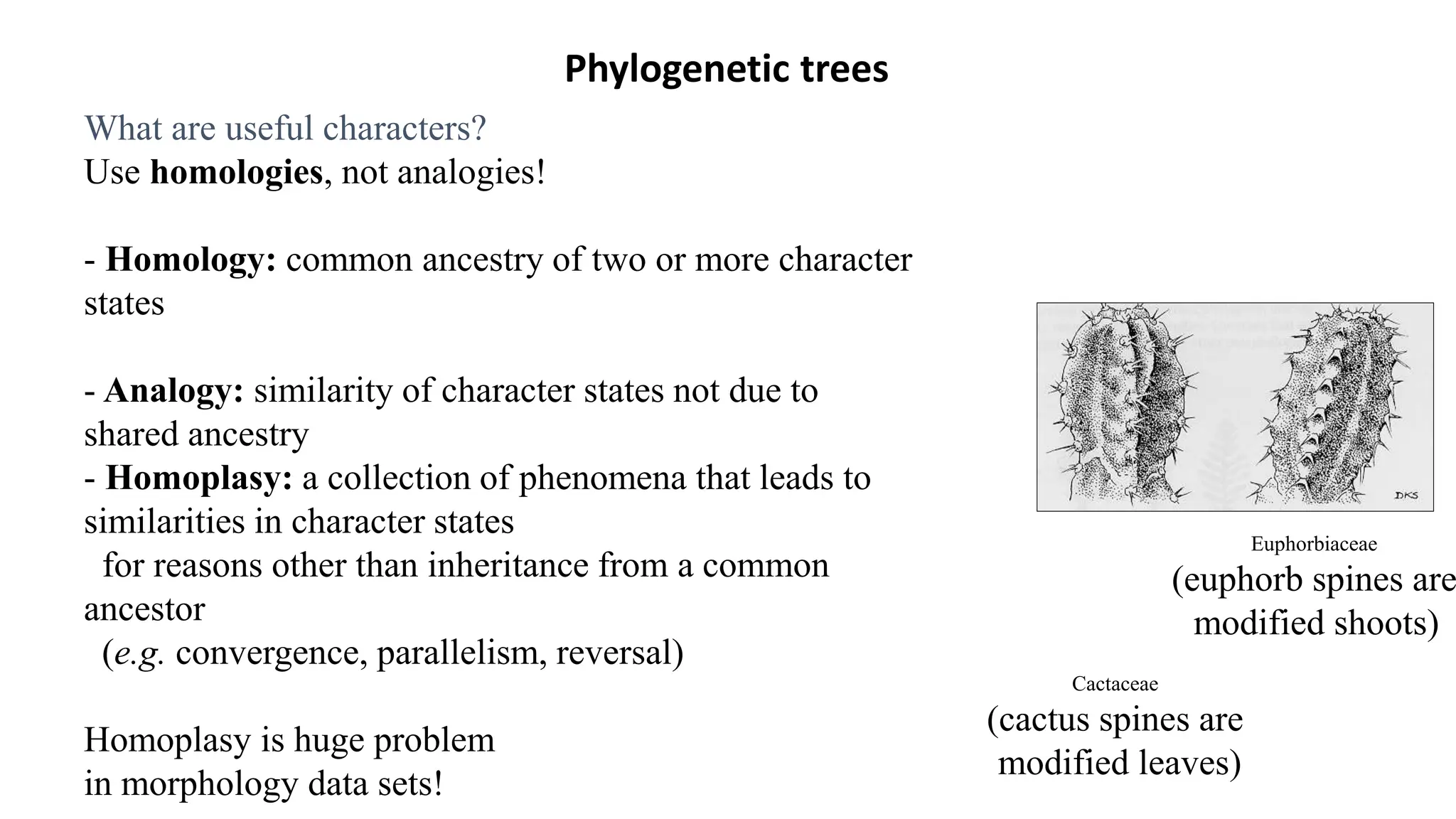
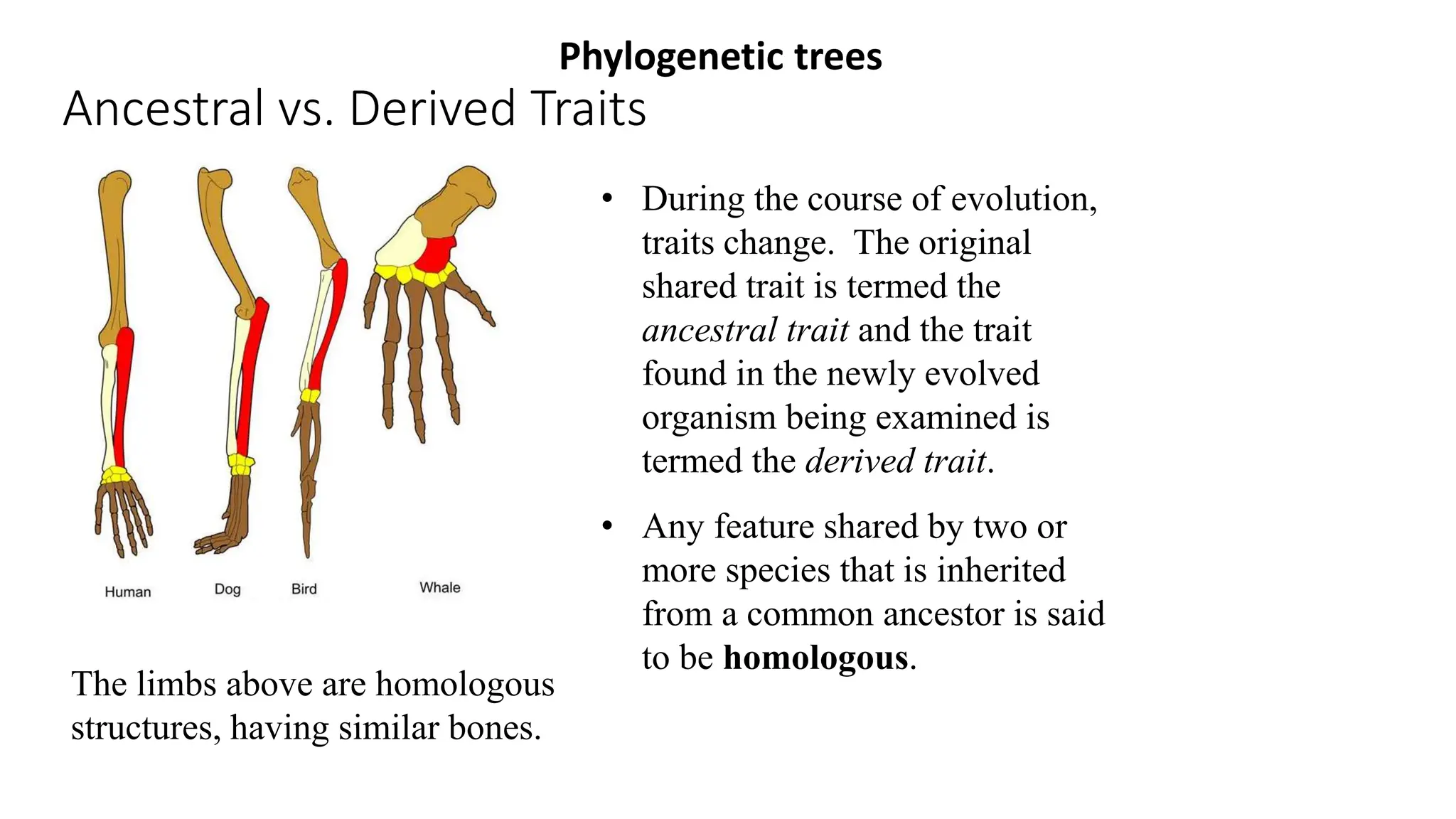
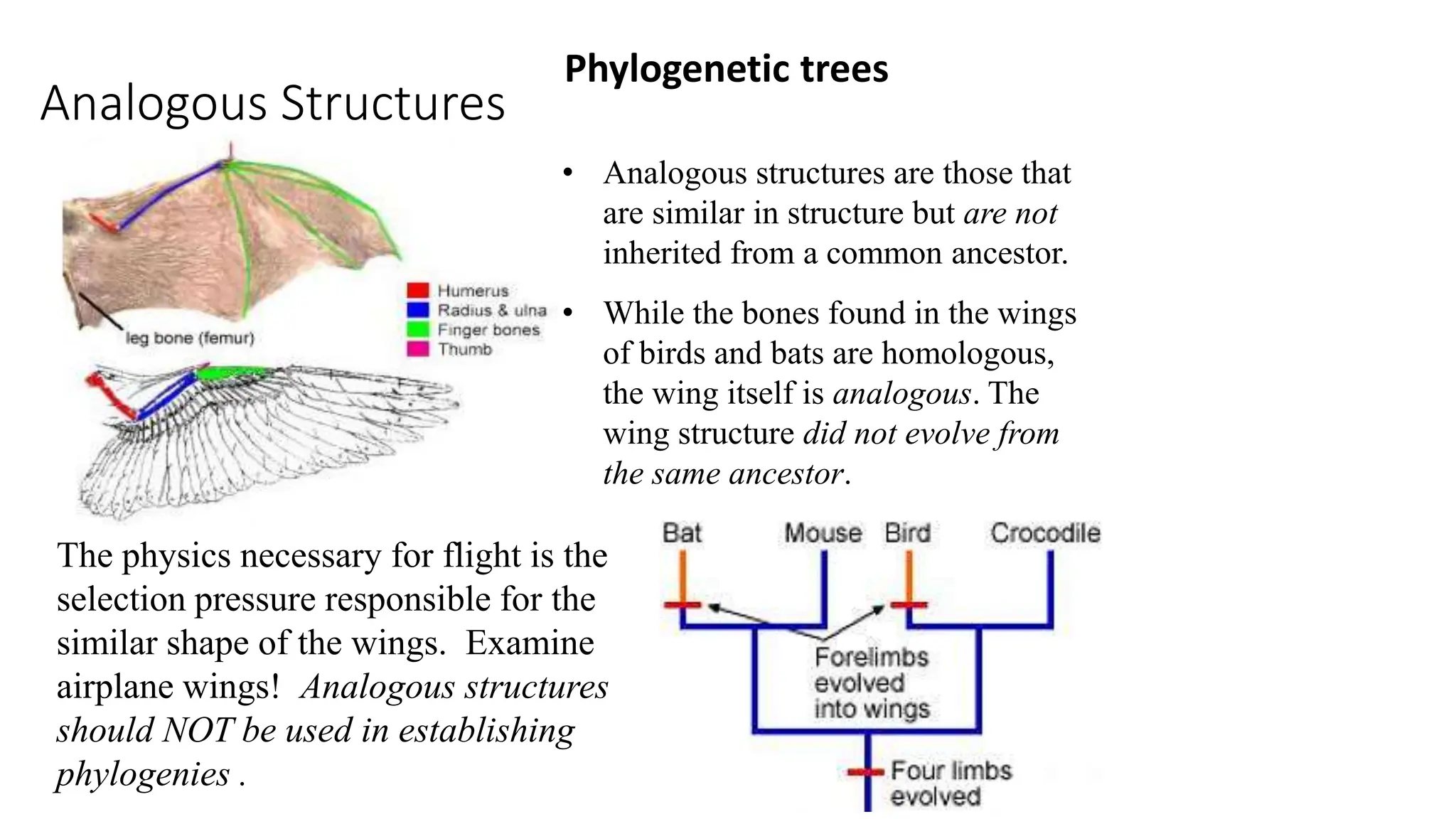
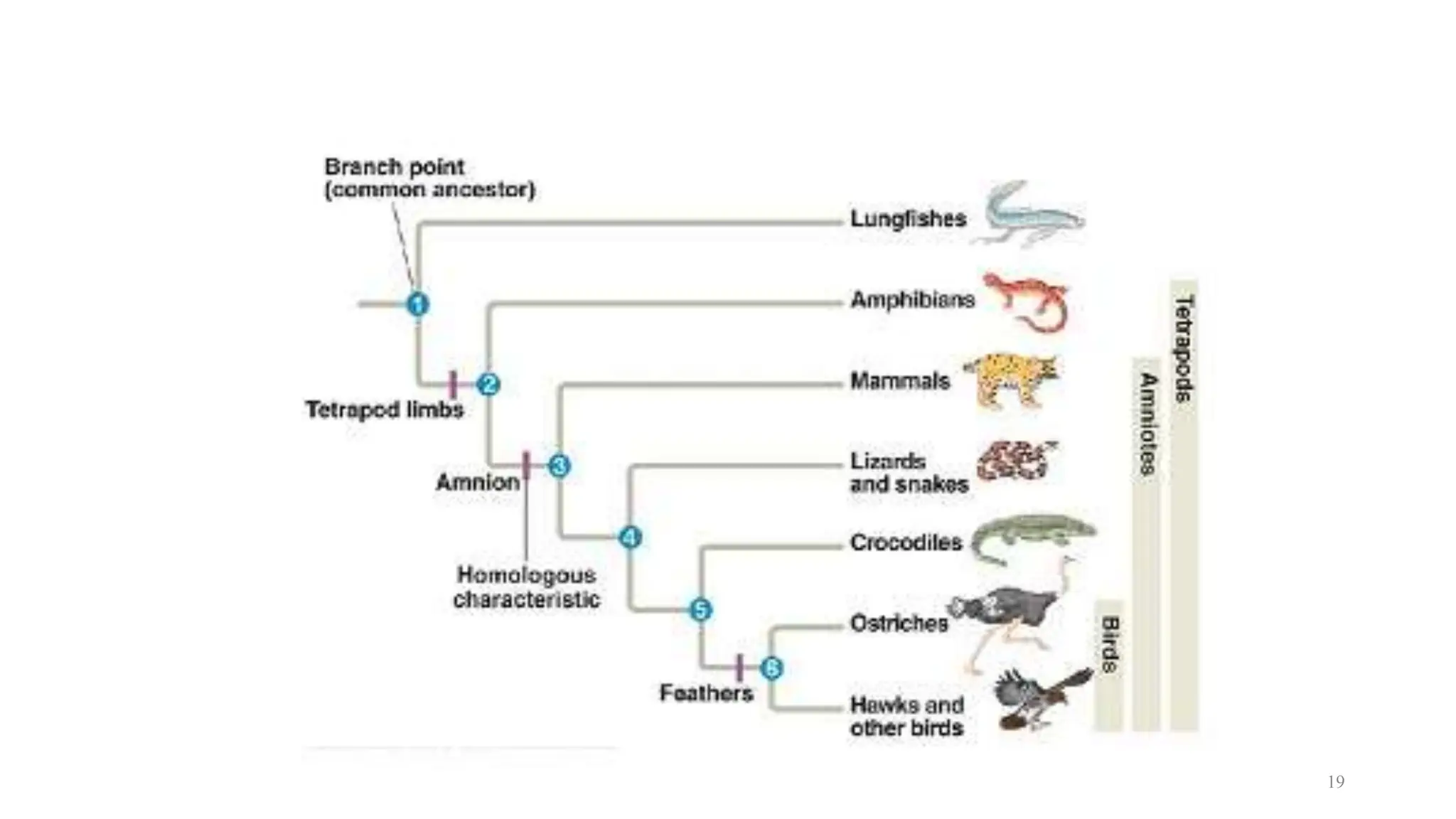
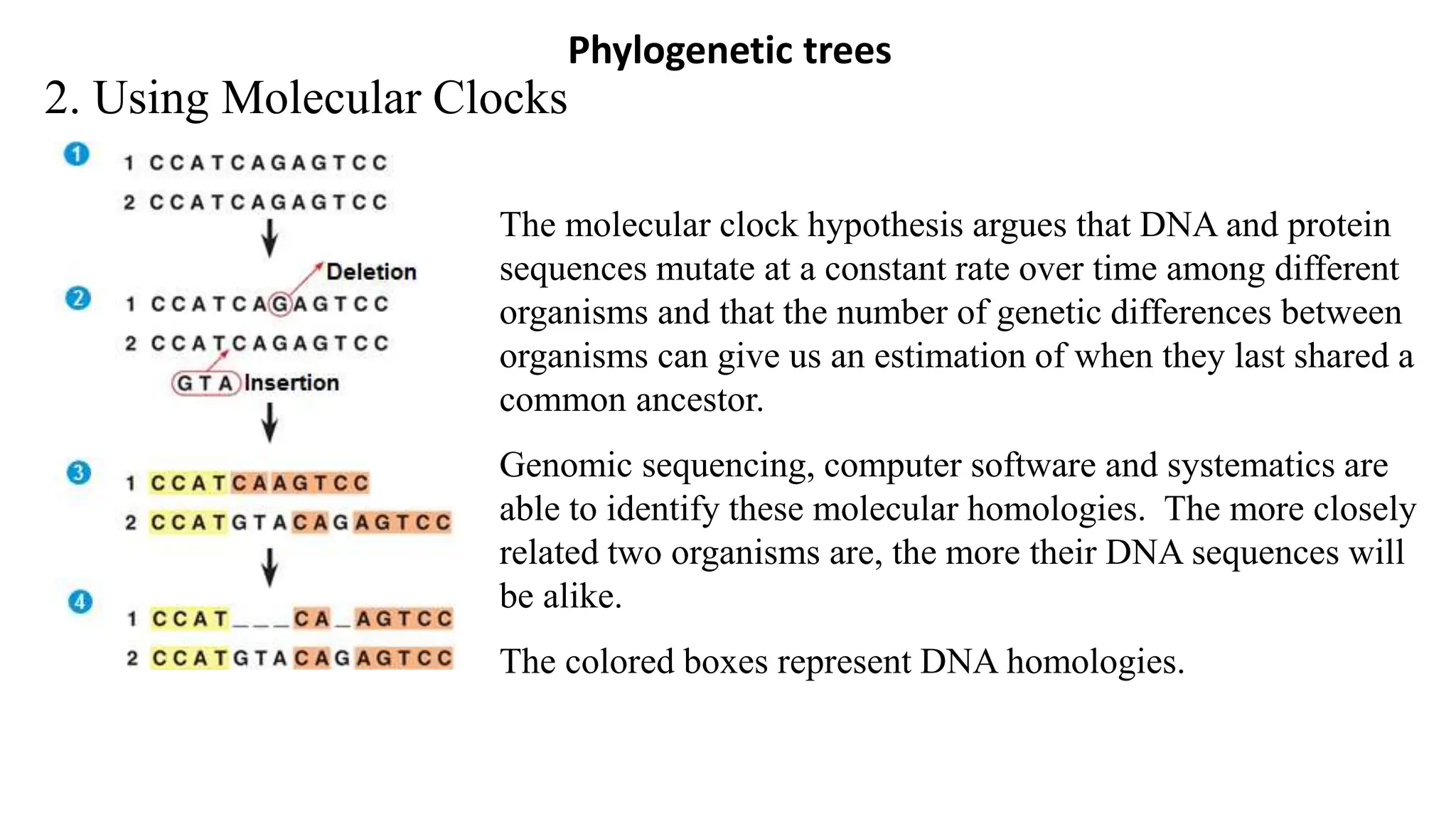
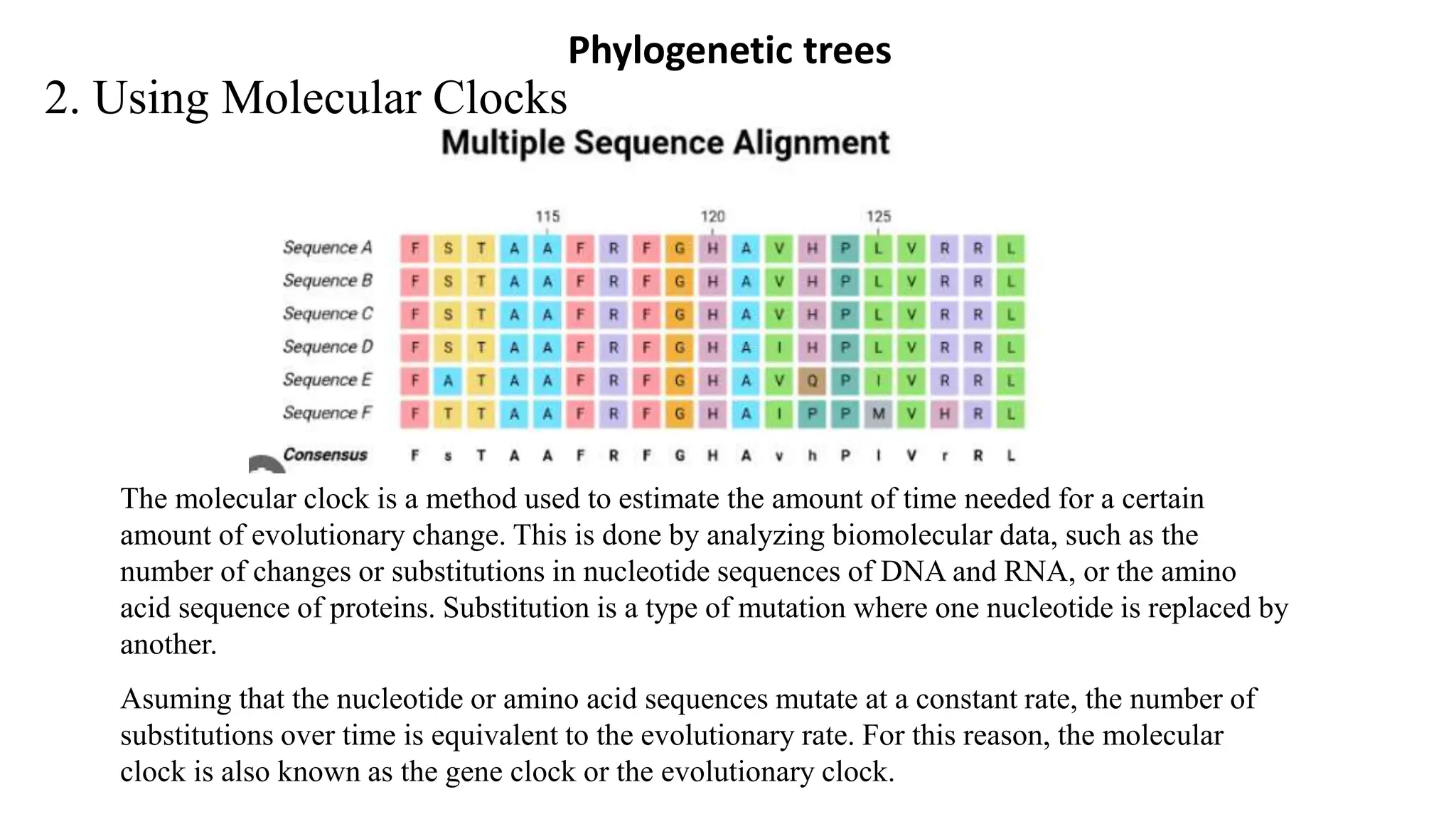
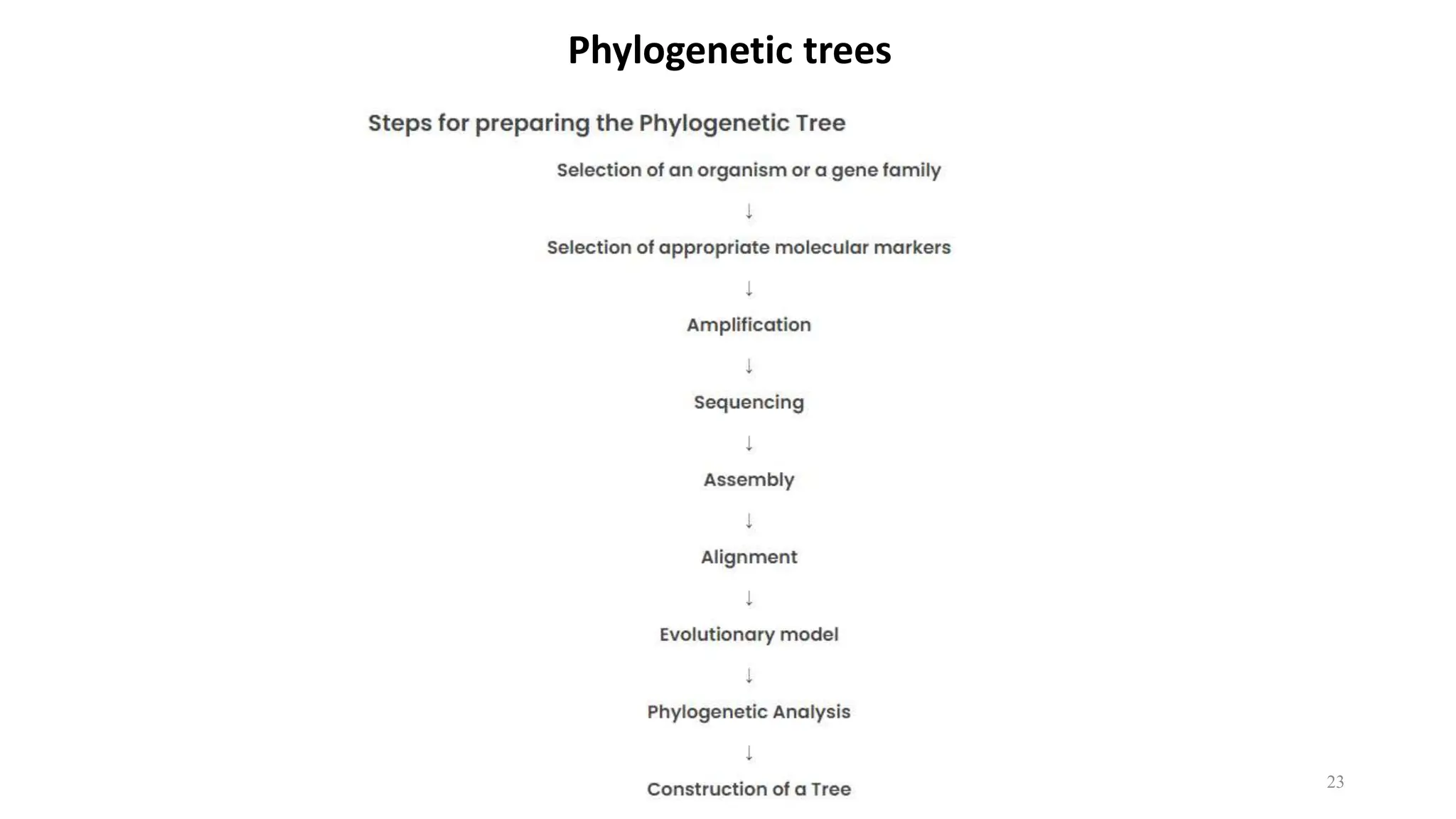
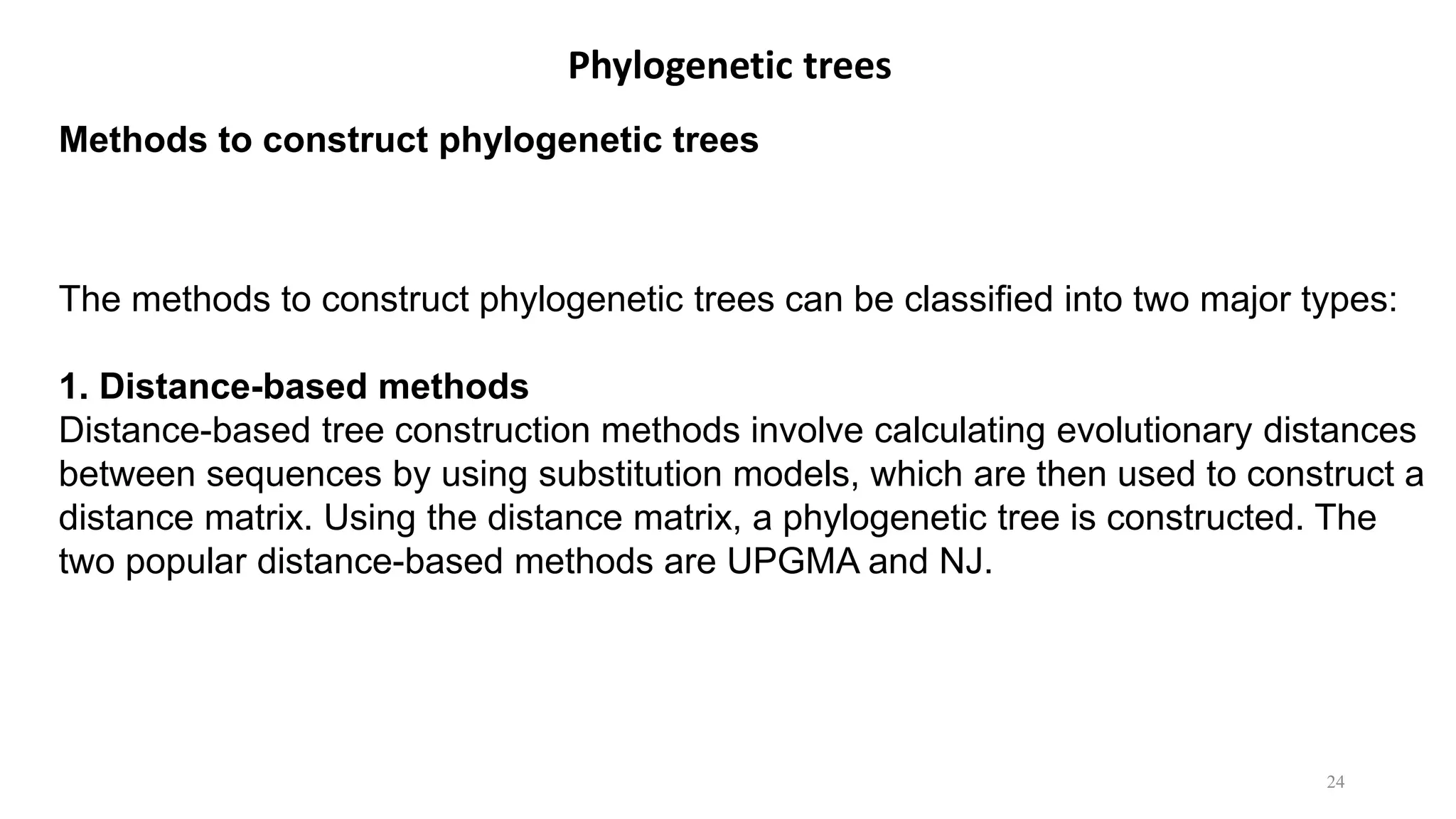
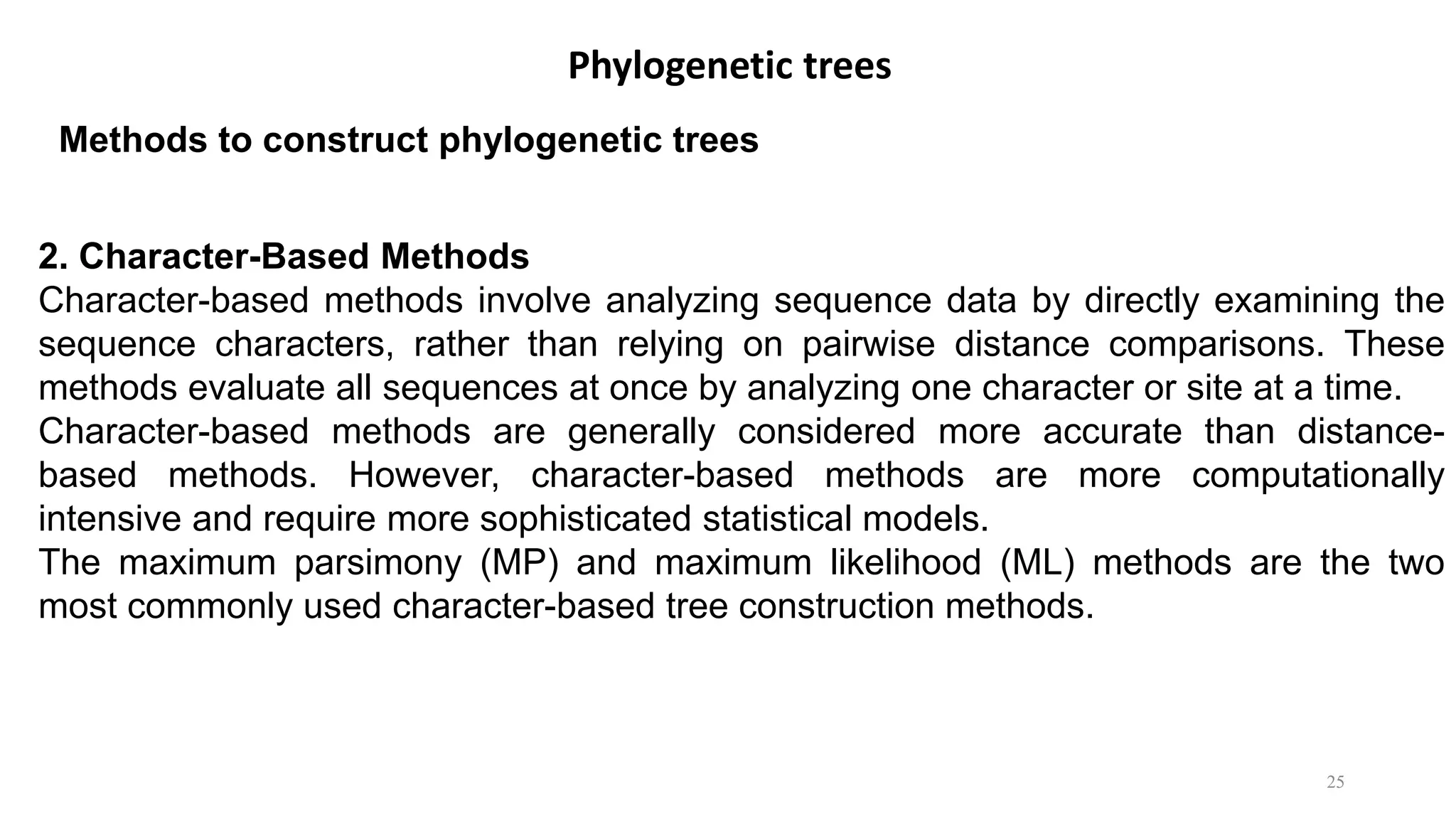
Scientists construct phylogenetic trees by analyzing both DNA sequences and protein sequences to identify molecular homologies that indicate evolutionary relationships. They use two main types of methods - distance-based methods that calculate evolutionary distances between sequences to build trees, and character-based methods that directly examine sequence characters to evaluate relationships. Specific genes, as well as whole genomes, can provide data for building phylogenetic trees through these computational analysis methods.