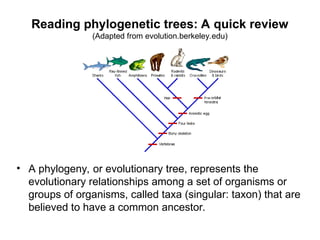
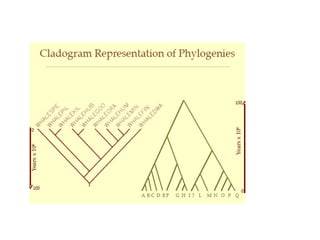
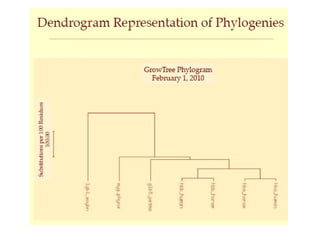
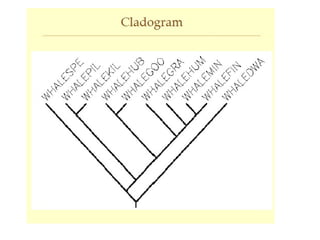
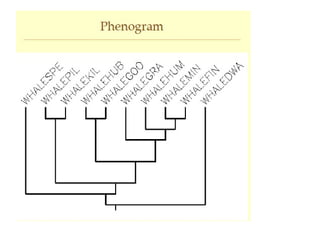
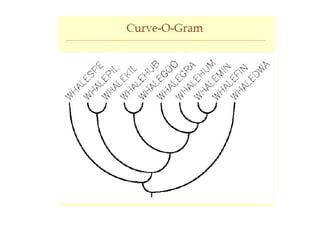
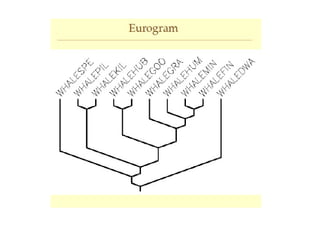
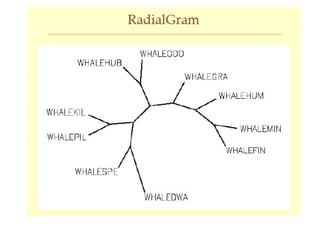
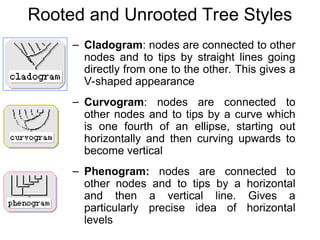
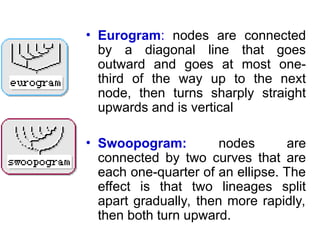
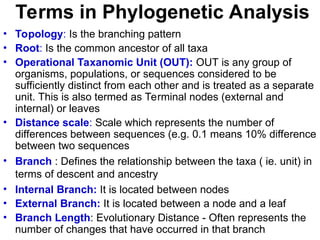
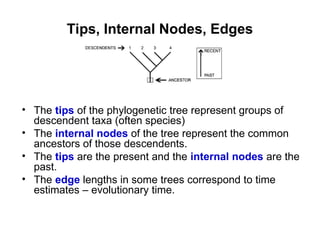
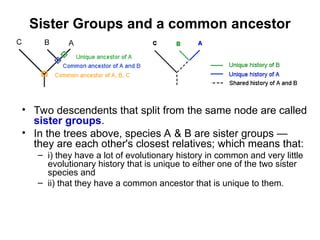
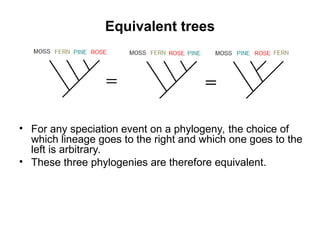
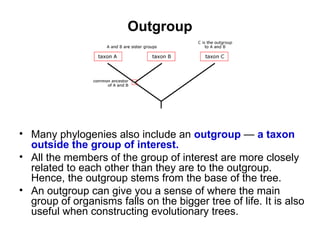
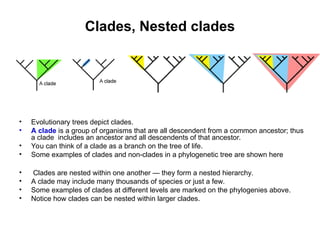
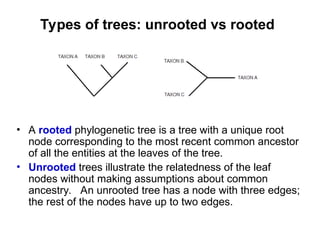
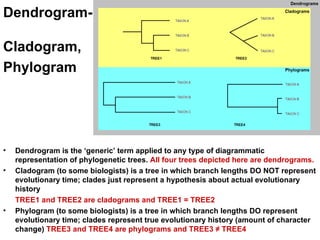
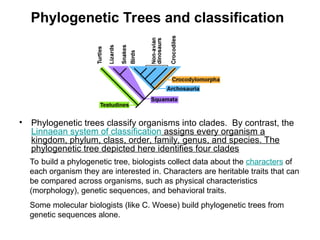
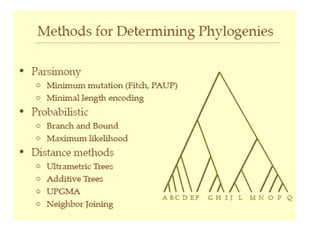
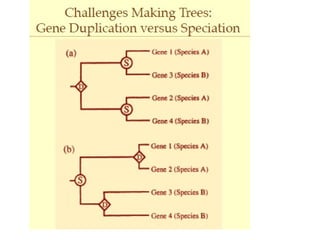
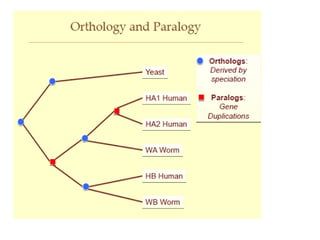
Phylogenetic trees are diagrams that depict evolutionary relationships among taxa believed to share a common ancestor, with various styles including cladograms, curvograms, and phenograms. Key terms in phylogenetic analysis include topology, branches, and clades, which help represent the relationships, ancestry, and characteristics of organisms. The document also distinguishes between rooted and unrooted trees and discusses the importance of outgroups and the classification of organisms based on evolutionary history.