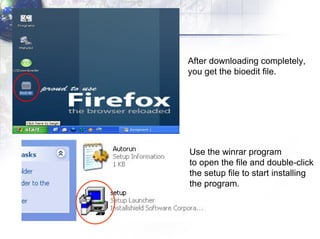
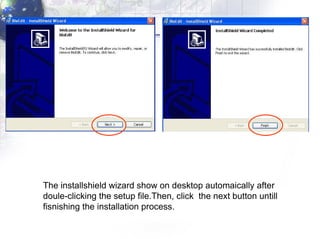
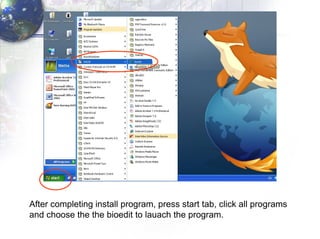
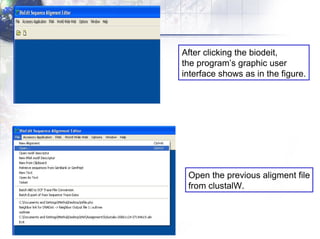
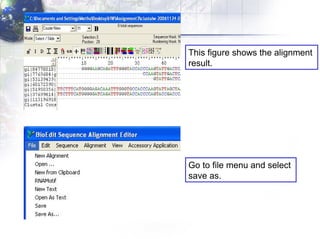
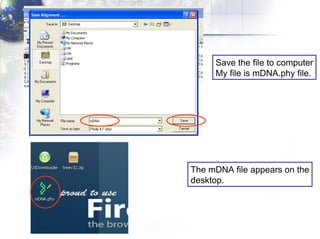
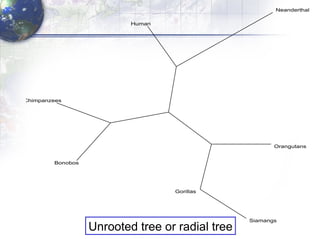
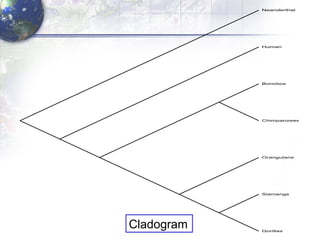
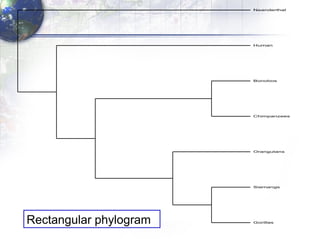
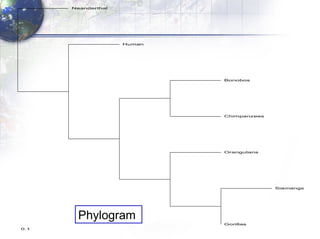
The document describes the process of constructing a phylogenetic tree from mitochondrial DNA sequence data retrieved from online databases. It involves the following steps:
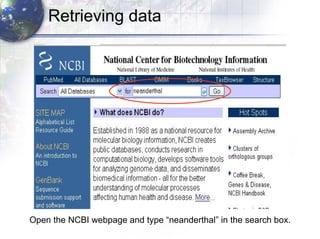
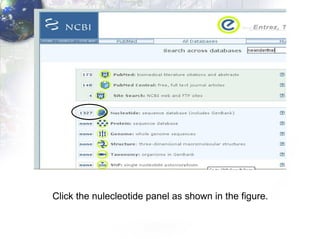
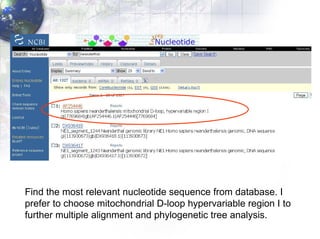
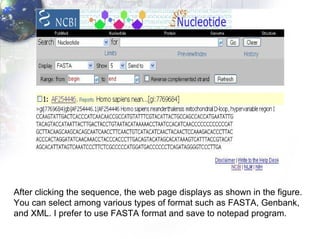
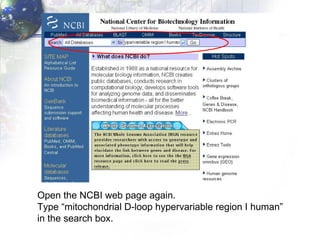
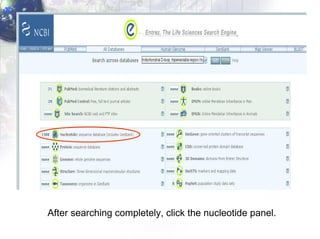
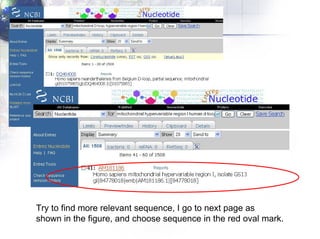
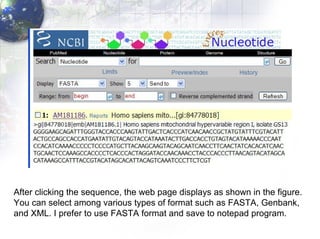
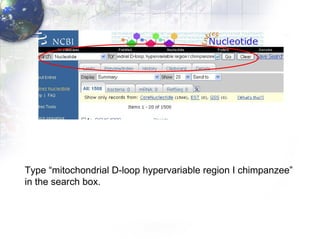
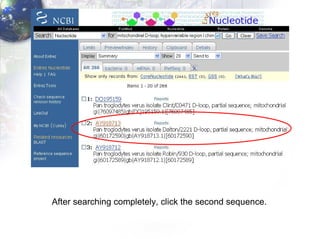
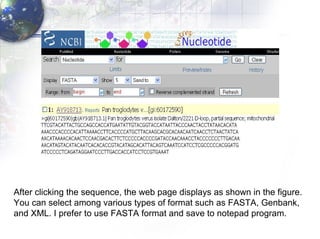
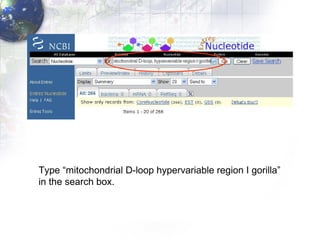
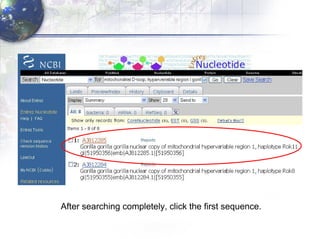
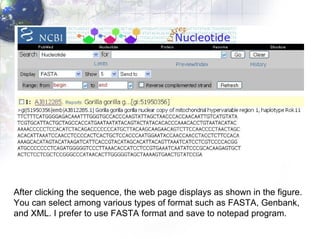
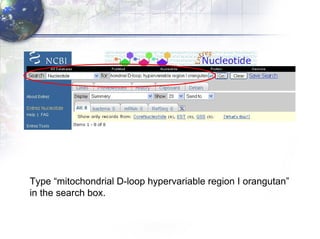
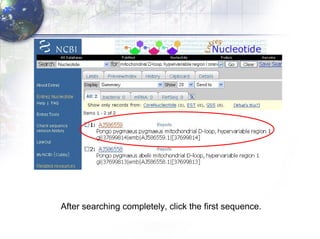
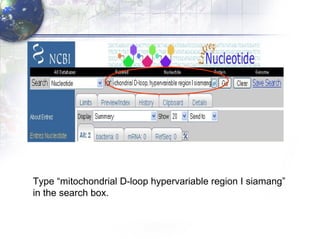
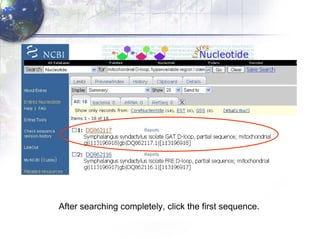
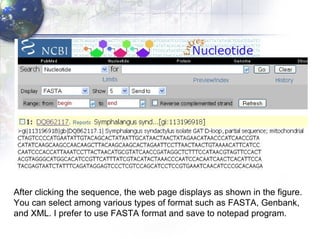
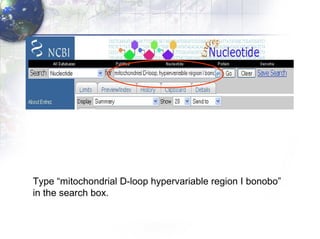
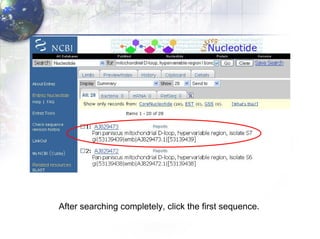
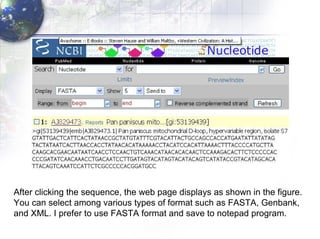
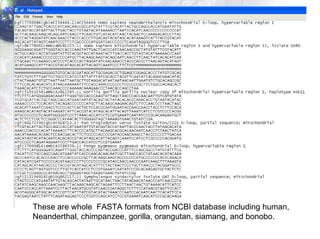
1) Retrieving sequence data from NCBI for humans, Neanderthals, chimpanzees, gorillas and other primates.
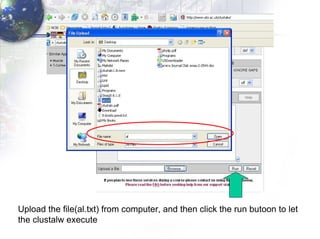
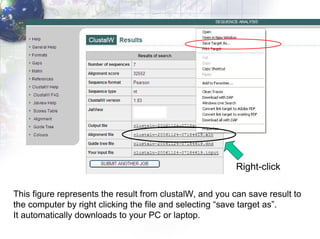
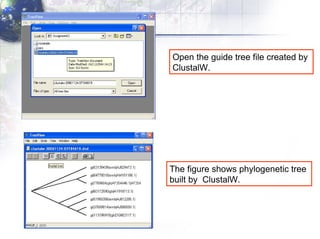
2) Aligning the sequences using ClustalW and viewing the alignment.
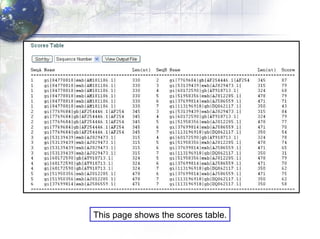
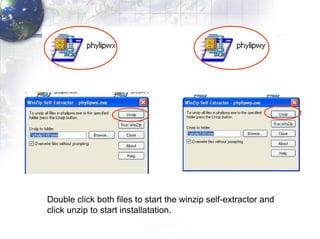
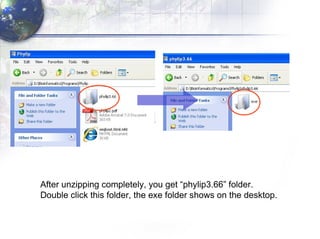
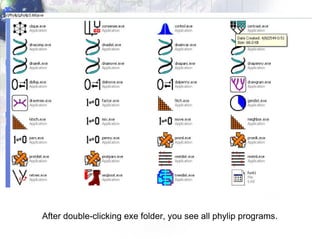
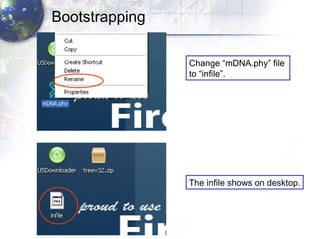
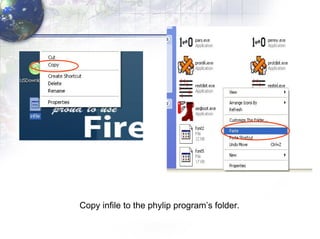
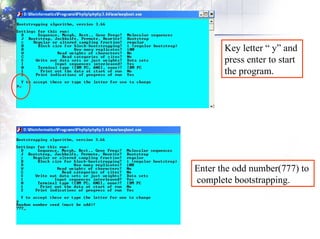
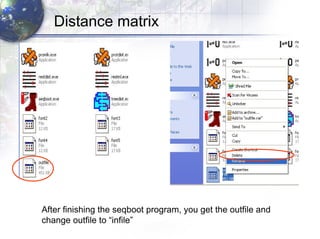
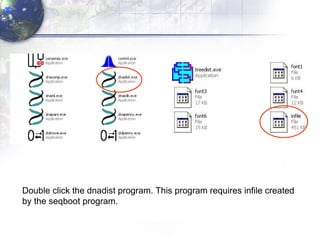
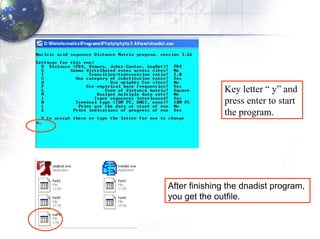
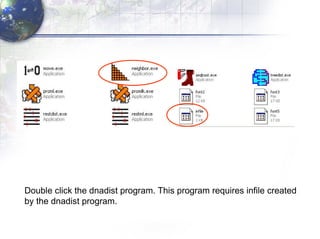
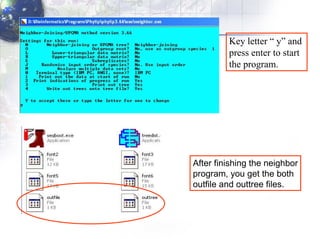
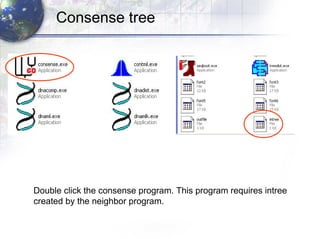
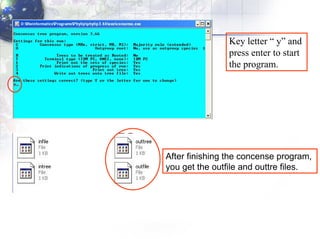
3) Using programs from the Phylip package like Seqboot, Dnadist, Neighbor and Consense to generate bootstrap trees, distance matrices and consensus trees.
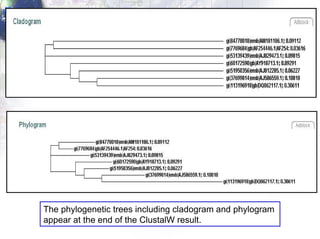
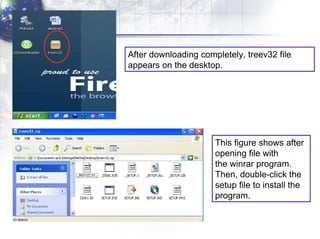
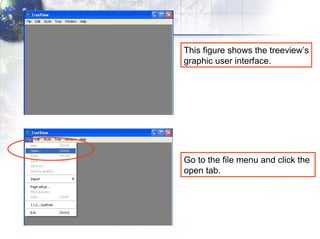
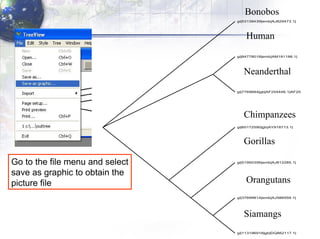
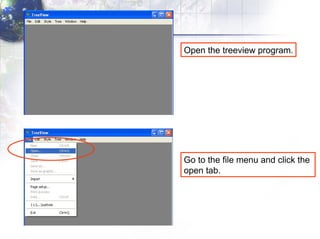
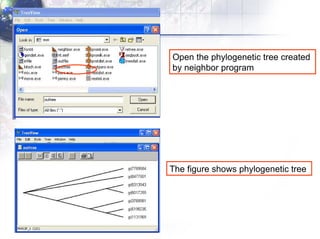
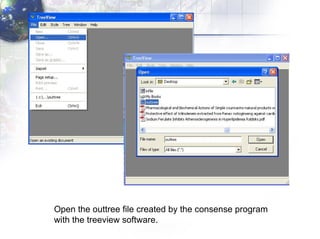
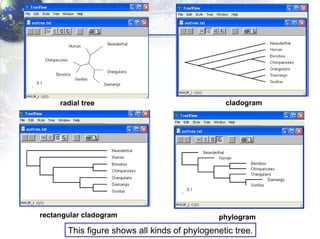
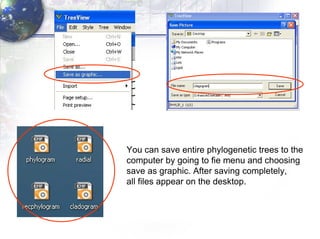
4) Viewing and saving the phylogenetic trees in TreeView.