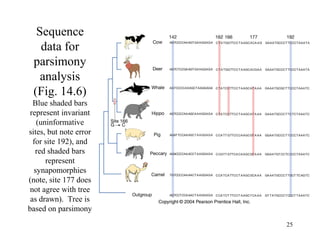
Phylogenetic trees reconstruct evolutionary relationships by grouping taxa with shared derived characteristics inherited from recent common ancestors. This document discusses methods for building phylogenetic trees, including cladistics which uses shared derived homologies (synapomorphies) to determine relationships. It also examines evidence for the evolutionary relationships of whales. Molecular studies of transposable elements and additional fossil evidence support whales evolving from artiodactyl ancestors, rather than being the sister group to artiodactyls.









![12
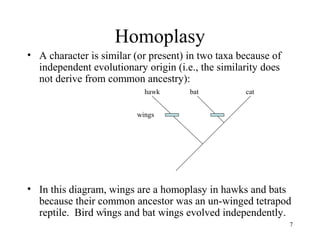
Two kinds of homology – 2
• Shared derived homology — a trait found in some
members of a group for which we are making a
phylogenetic tree (and which was NOT present in the
common ancestor of the entire group) — synapomorphy
– For example: hair is (potentially) a shared derived homology in the
group [dogs, humans, lizards]
– Synapomorphies DO provide phylogenetic information about
relationships within the group being studied
– In this particular case, if hair is a synapomorphy in dogs and
humans, then dogs and humans share a common ancestor that is
not shared with lizards, and the common dog-human ancestor must
have lived more recently than the common ancestor of all three
taxa](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-12-320.jpg)
![13
A tree for [dogs, humans, lizards] – 1
lizard human dog
hair
backbone
• The TWO major assumptions that we are making
when we build this tree are:
1) hair is homologous in humans and dogs
2) hair is a derived trait within tetrapods](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-13-320.jpg)
![14
A tree for [dogs, humans, lizards] – 2
lizard human dog
hair
backbone
• In the absence of other information, the assumption of
homology of hair in humans and dogs is justified by
parsimony (fewest number of evolutionary steps is
most likely = simplest explanation)
• Also we can check to see that hair is formed in the
same way by the same kinds of cells, etc.](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-14-320.jpg)
![15
A tree for [dogs, humans, lizards] – 3
• These trees (in which hair is considered a homoplasy
in dogs and humans) are less parsimonious than the
one on the previous slide, because they require two
independent evolutionary origins of hair
human lizard dog
hair
backbone
hair
dog lizard human
hair
backbone
hair](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-15-320.jpg)


![18
Outgroups – 2
• In the present example, [dog, human, lizard] are
all amniote tetrapods. The anamniote tetrapods
(amphibia) make a reasonable outgroup for this
problem
• No amphibia have hair, therefore absence of hair
[amphibia, lizards] is primitive (plesiomorphic)
and presence of hair [dogs, humans] is derived
(apomorphic)
• So, presence of hair is a shared derived character
(synapomorphy), and dogs and humans are more
closely related to each other than either is to
lizards](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-18-320.jpg)
![19
A tree for [dogs, humans, lizards] – 4
• The presence of hair is apomorphic (derived) because
no amphibians have hair
lizard human dog
hair
backbone
Amphibia
amniotic egg](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-19-320.jpg)

![21
A tree for [dogs, humans, lizards] – 5
• Tree (a) is most parsimonious, so we’ll take that as our best
estimate of the true phylogeny of [dog, human, lizard]
• Of course, if we studied different characters, or used a different
outgroup, our phylogenetic tree could change
human lizard dog
hair
backbone
hair
dog lizard human
hair
backbone
hair
lizard human dog
hair
backbone
(a)
(b)
(c)](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-21-320.jpg)

![23
The
Artiodactlya
hypothesis
for the
evolutionary
relationships
of Cetacea
(Fig. 14.4 a)
Odd-toed
ungulates
(Perissodactyla
[horses, rhinos])
are the outgroup](https://image.slidesharecdn.com/phylogeny-140802005657-phpapp01/85/Phylogeny-23-320.jpg)