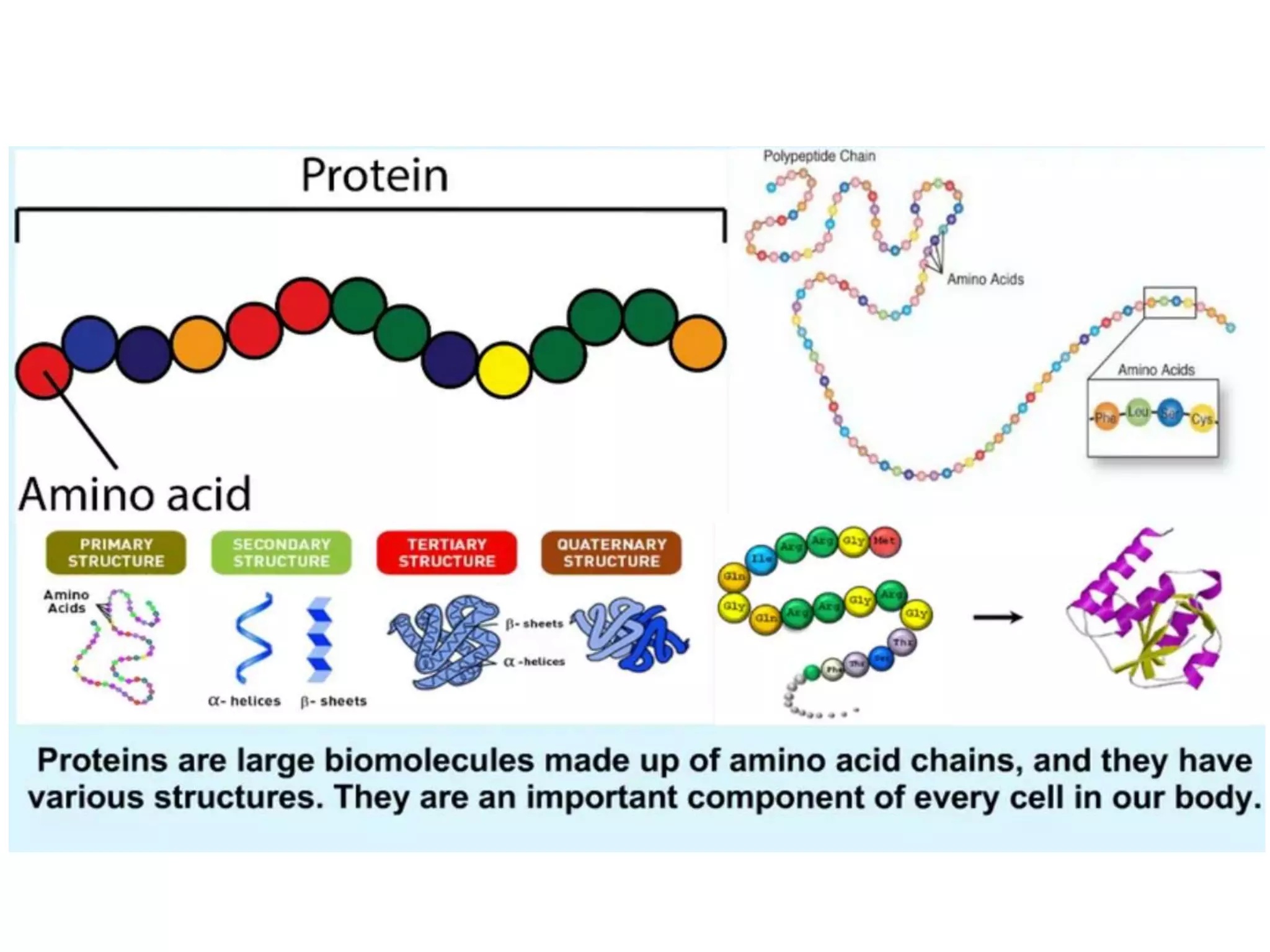
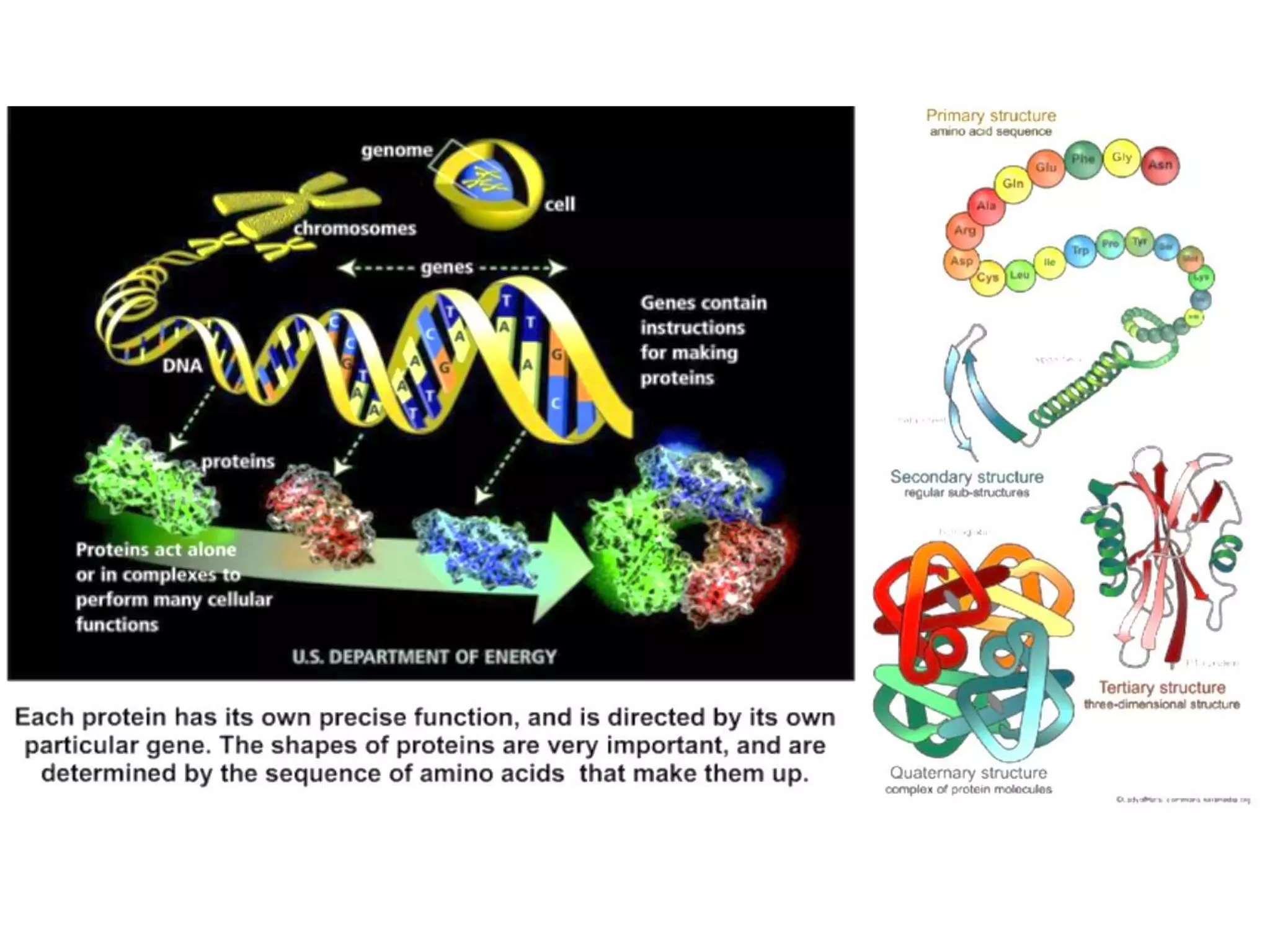
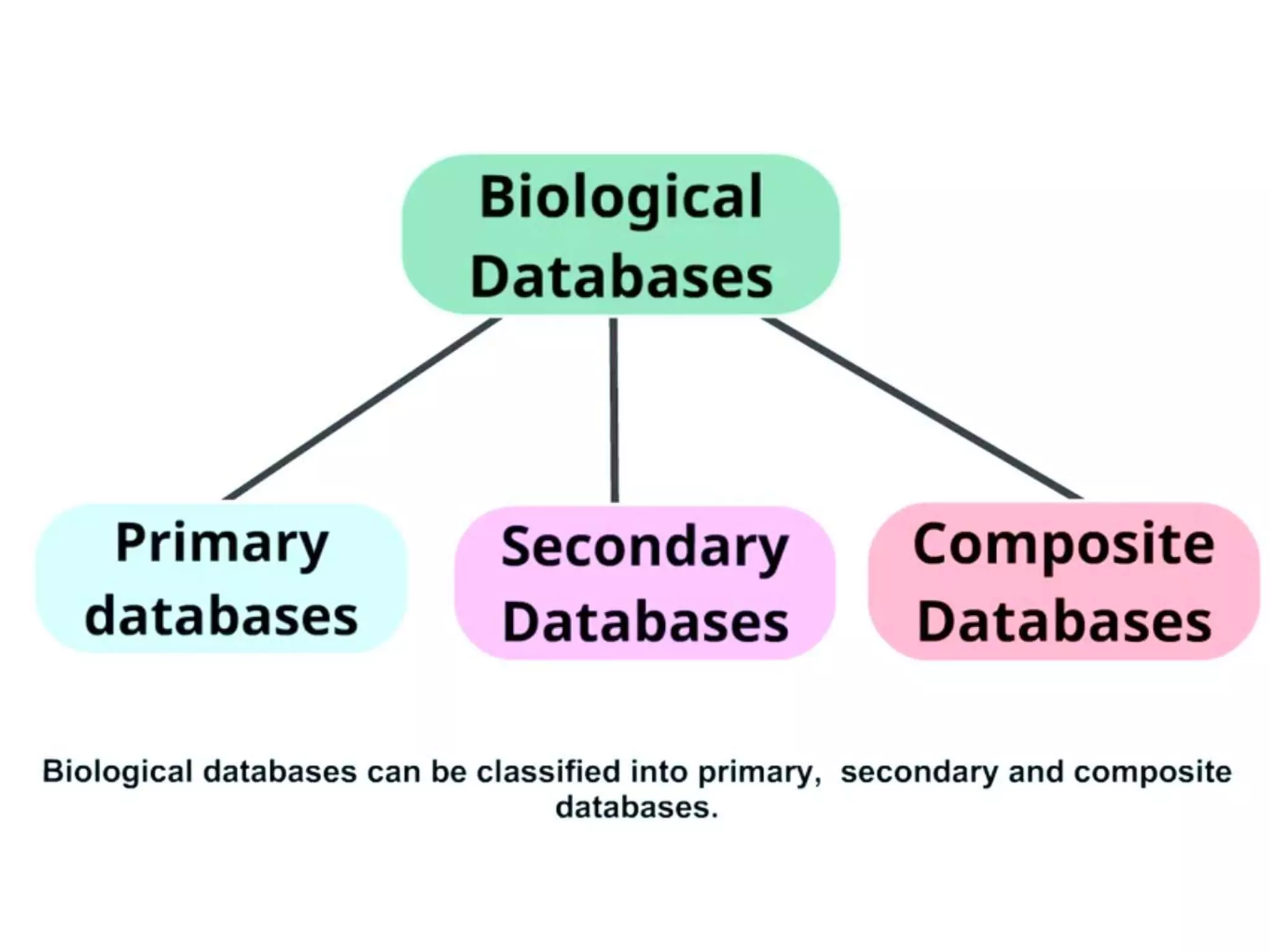
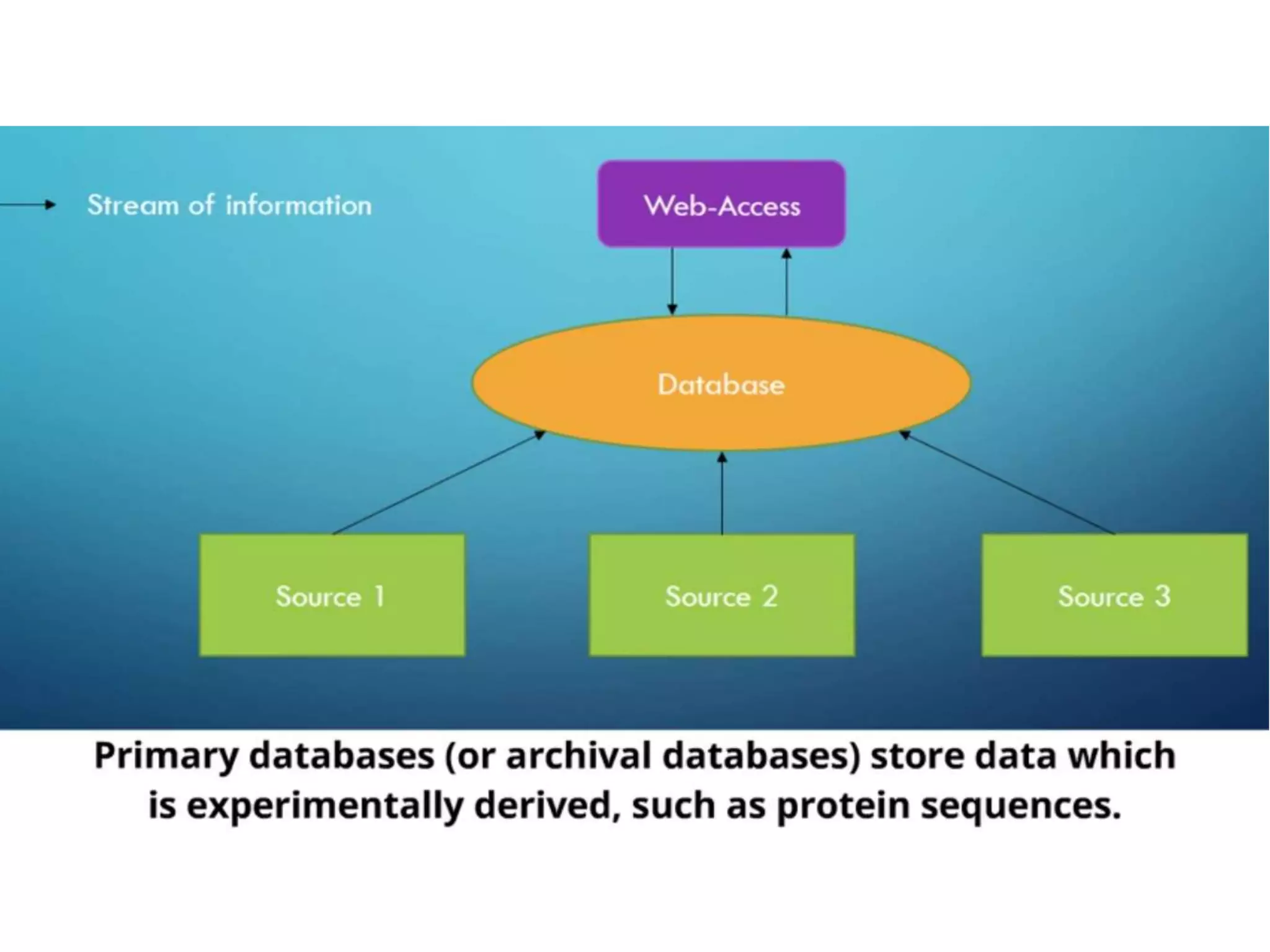
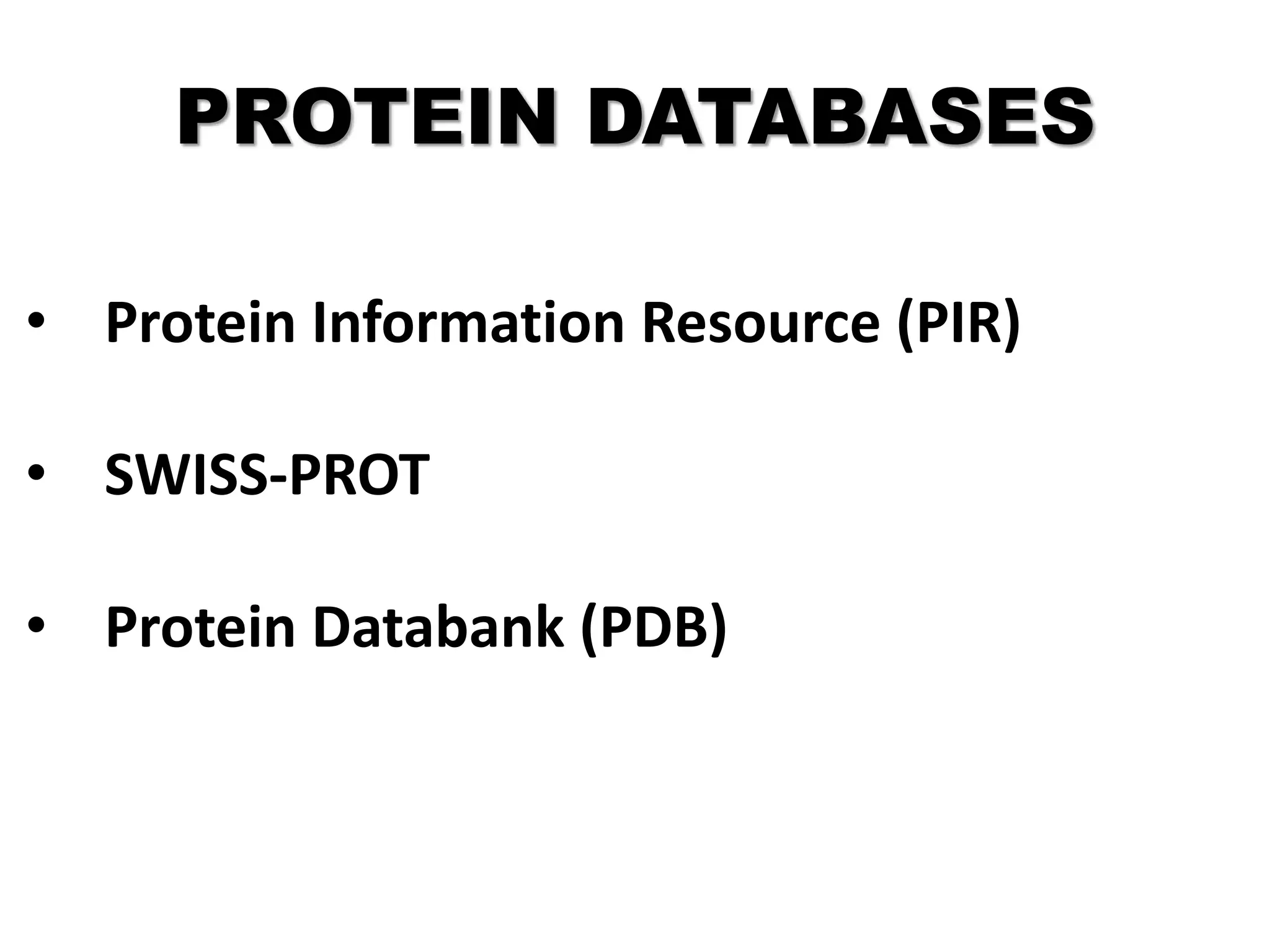
Protein databases contain information on protein sequences, structures, and functions. The major protein databases are:
- Protein Data Bank (PDB) which contains 3D protein structures determined via X-ray crystallography or NMR.
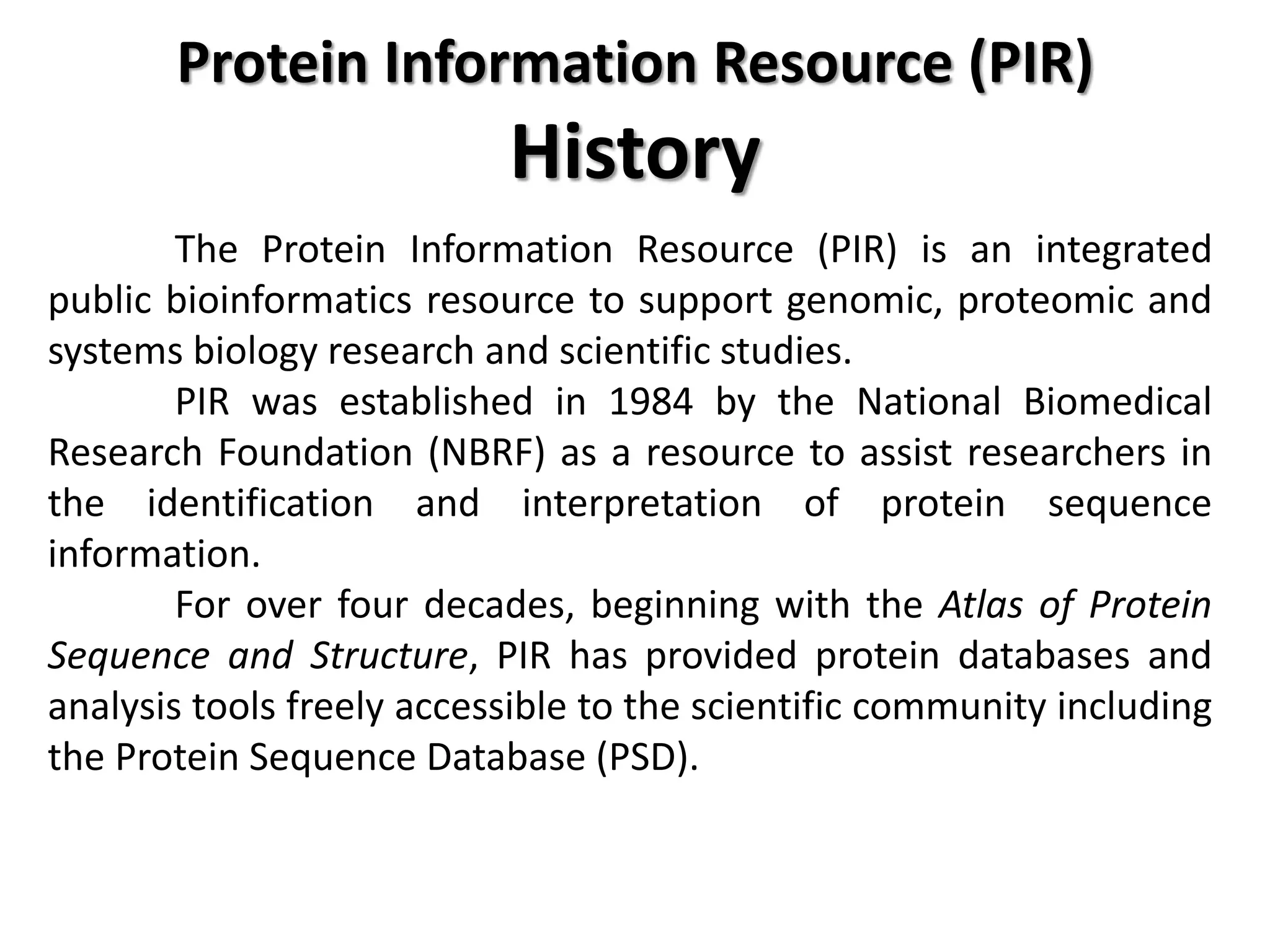
- Swiss-Prot which contains manually annotated protein sequences and functions.
- TrEMBL which supplements Swiss-Prot with automatically annotated translations of DNA sequences.
Protein databases are important for comparing proteins, understanding relationships between proteins, and aiding the study of new proteins. Searching databases is often the first step in protein research.