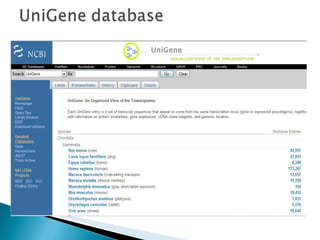
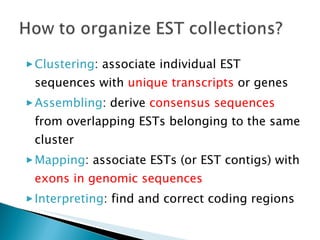
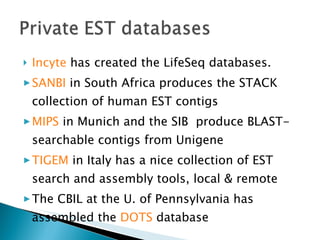
ESTs are short sequences of DNA that represent genes expressed in certain tissues or organisms. They provide a quick and inexpensive way for scientists to discover new genes and map their positions in genomes. ESTs represent a snapshot of genes expressed in a tissue at a given time. Sequencing the beginning or end of cDNA clones produces 5' and 3' ESTs, which can help identify genes and study gene expression and regulation.