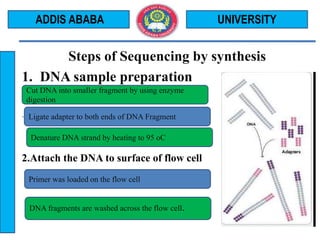
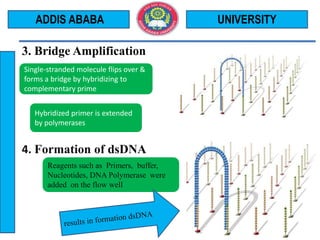
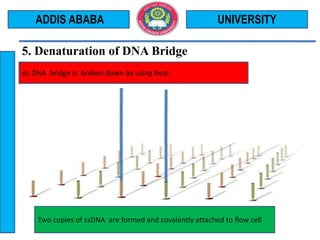
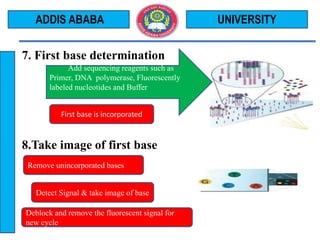
The document outlines the Illumina sequencing by synthesis method in 12 steps: 1) DNA sample preparation and attachment to a flow cell, 2) bridge amplification to clone the DNA fragments, 3) determination of the first base by addition of fluorescently labeled nucleotides and imaging, 4) repeating the process to determine the second and subsequent bases, 5) generating sequenced reads. The sequenced reads are then 6) aligned and analyzed by comparing to a reference sequence to identify differences.