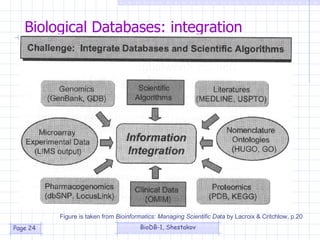
This document provides an overview of an introductory course on biological database systems. It discusses the course content, objectives, structure and readings. The course will cover database concepts, biological data representation, database design, and example integration systems. The goal is to provide basic knowledge of designing biological databases for molecular biology.