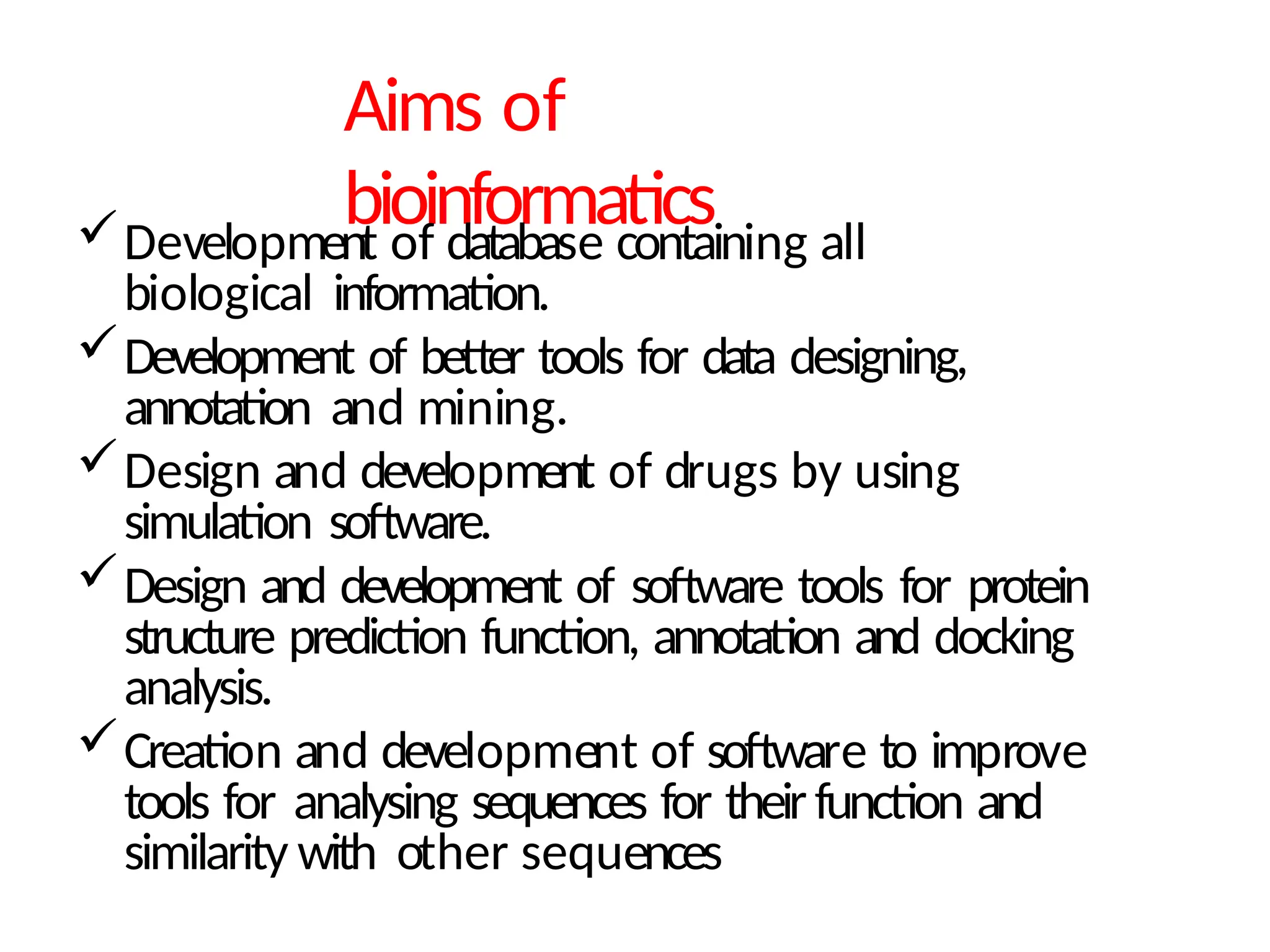
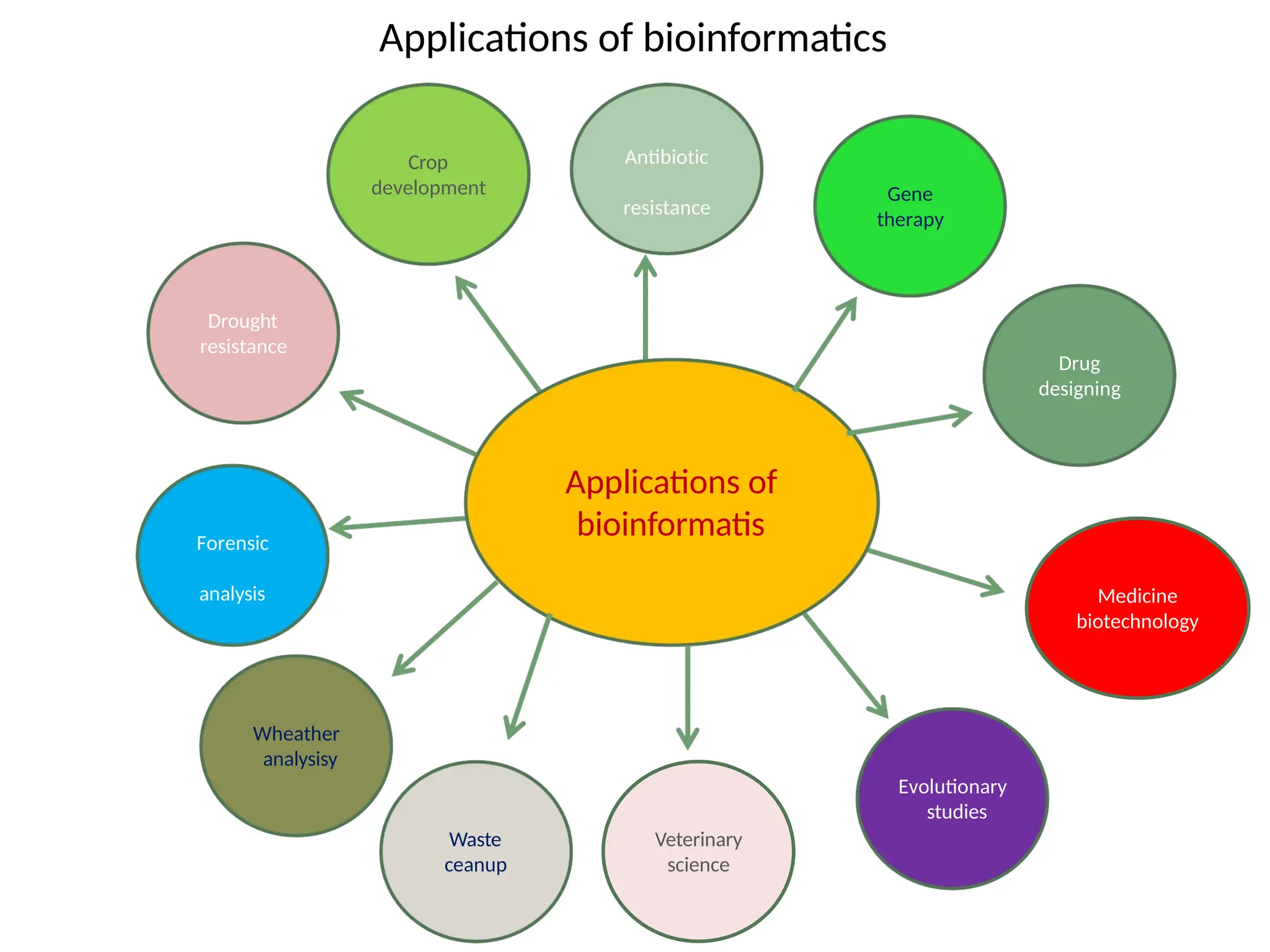
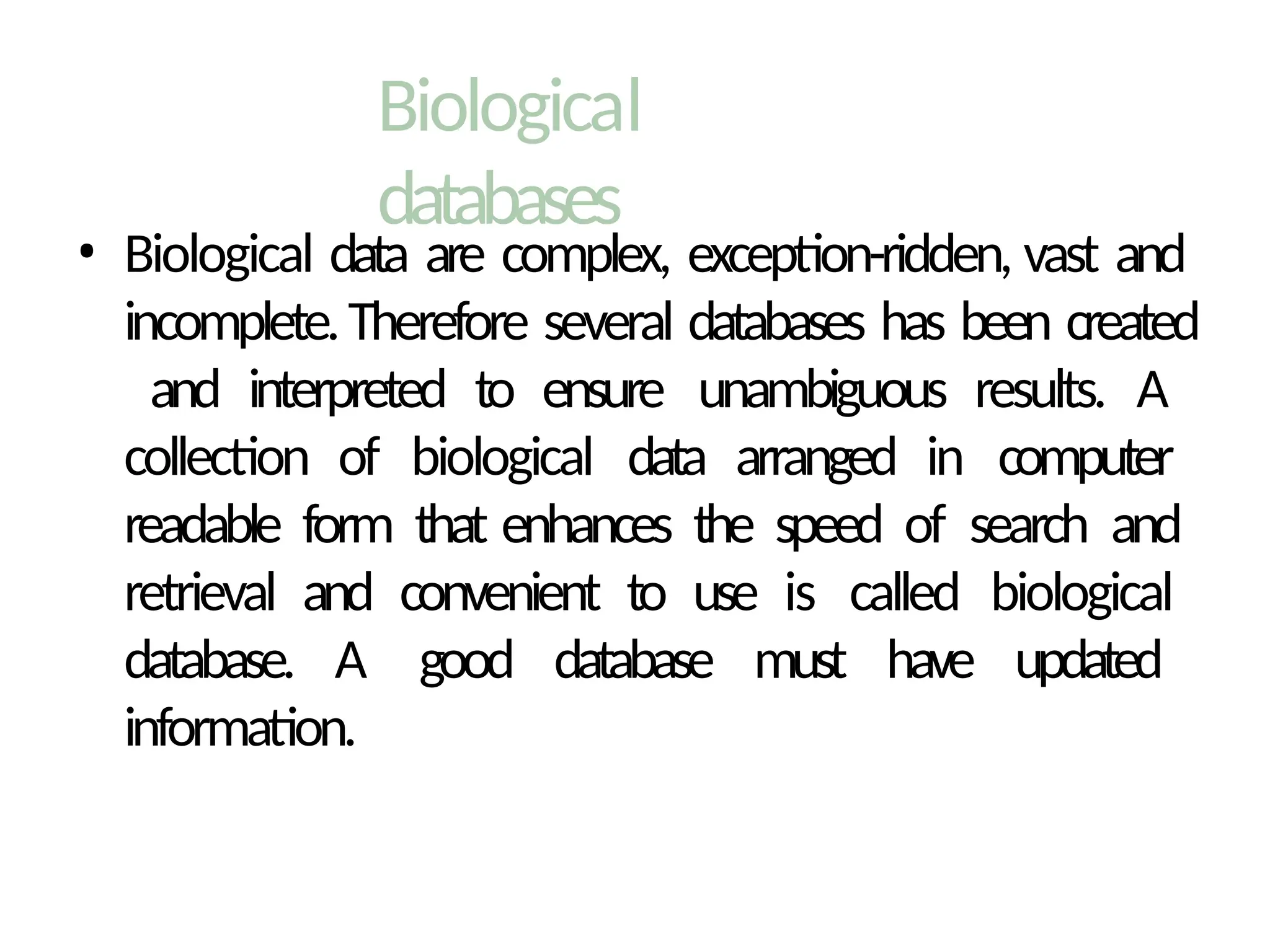
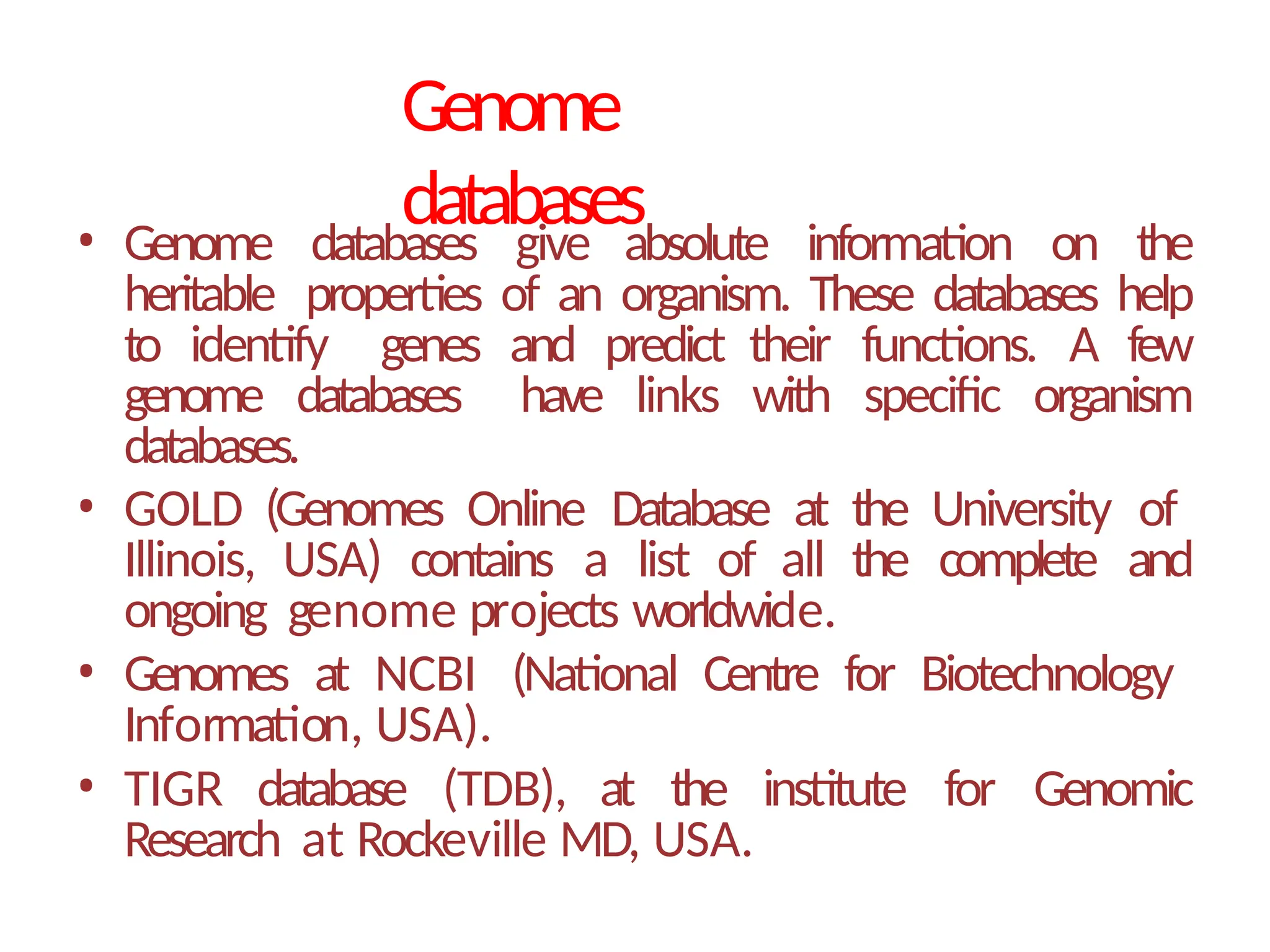
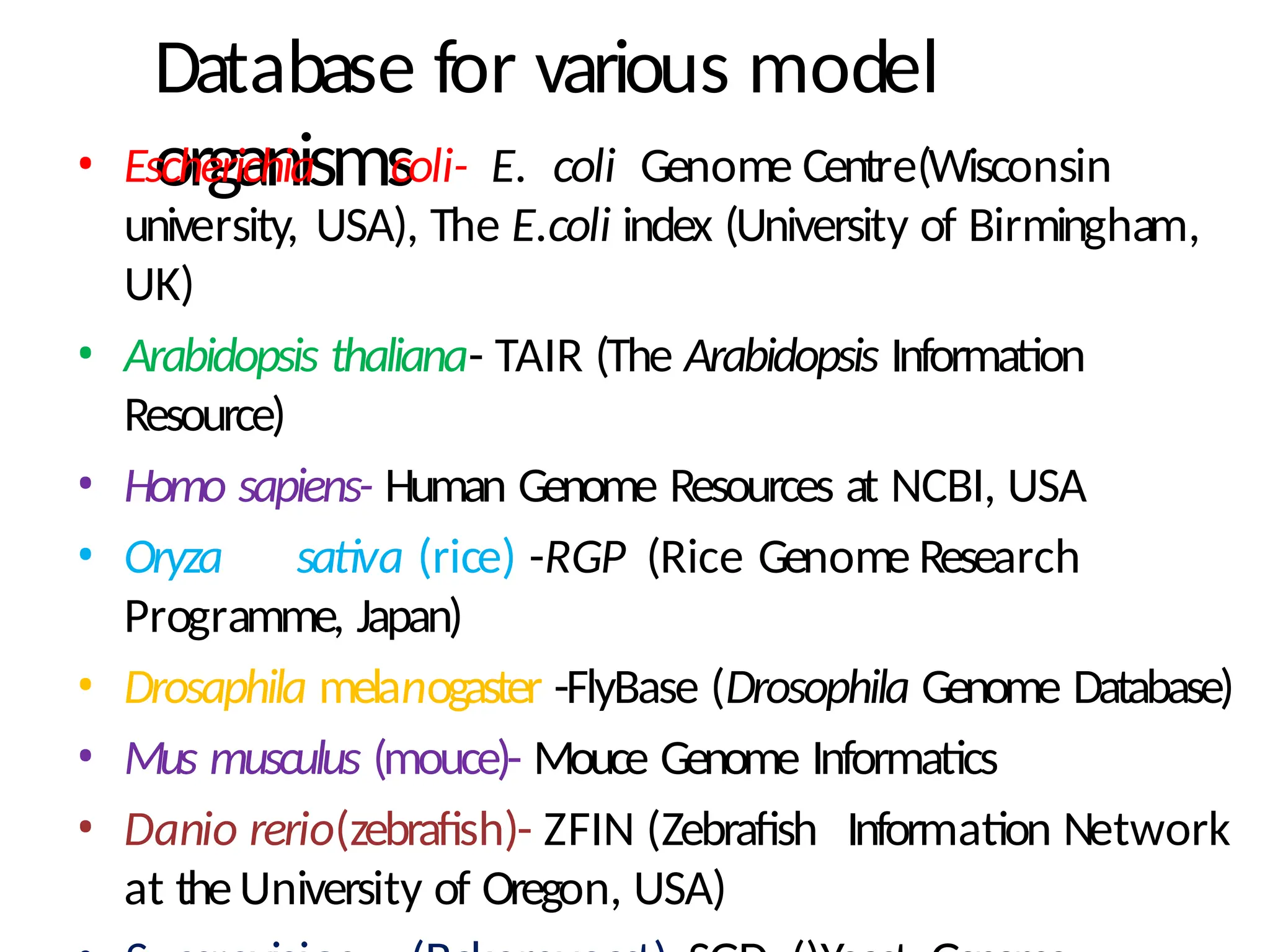
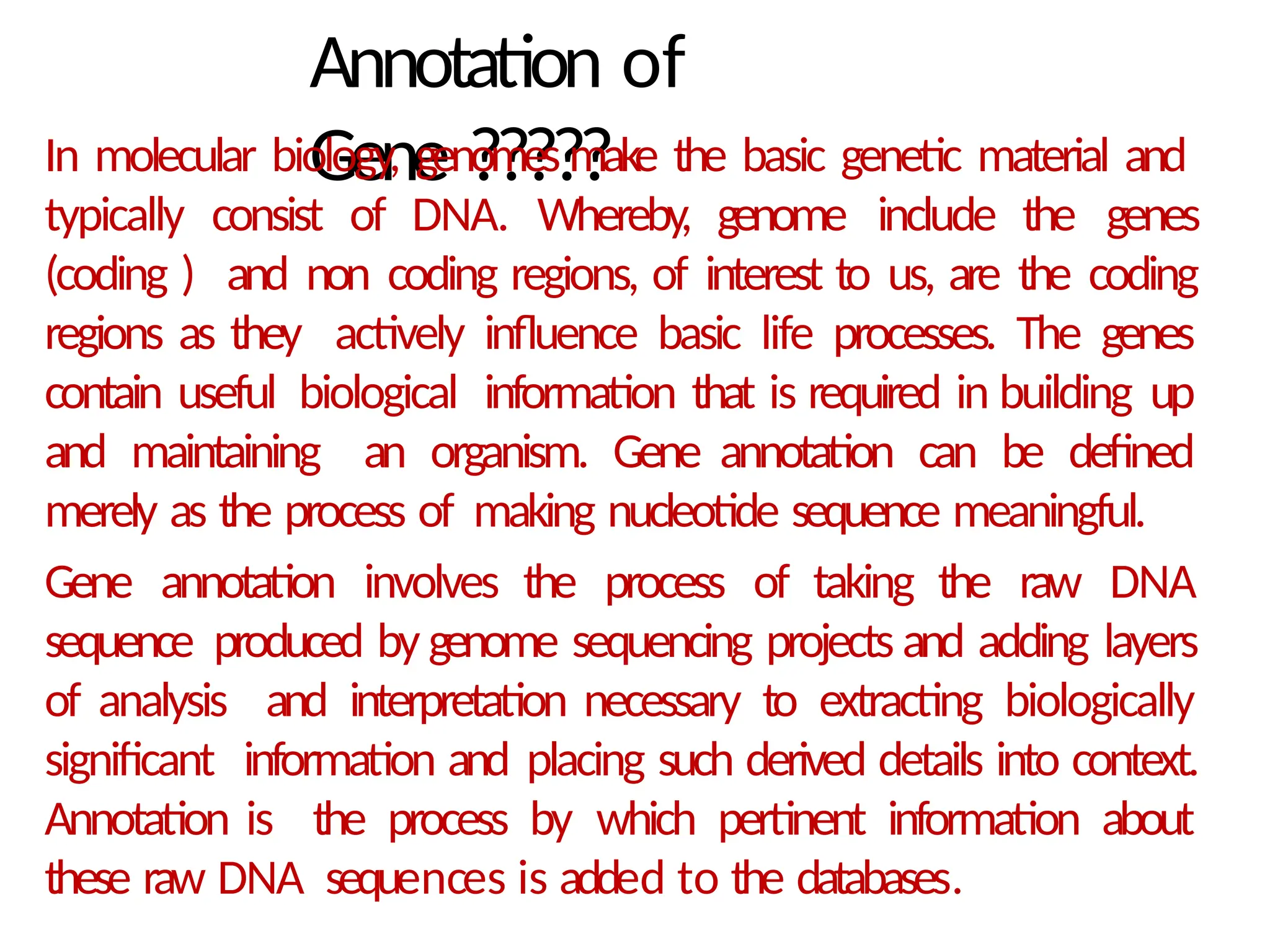
The document provides an introduction to bioinformatics, emphasizing its role in managing biological data through computational techniques, and outlines its applications, including drug design and gene therapy. It explains various types of biological databases, including nucleotide and protein structure databases, and their importance in storing and retrieving complex biological information. The document also covers gene annotation processes and the significance of accession numbers in identifying sequences within databases.