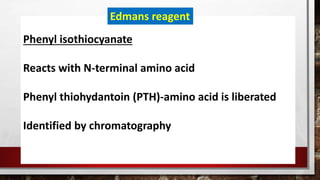
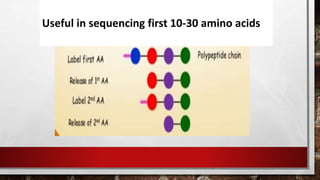
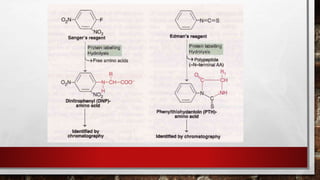
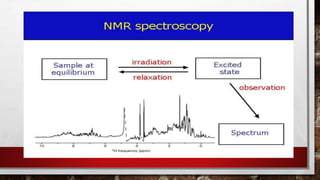
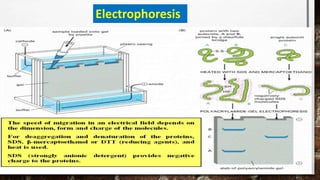
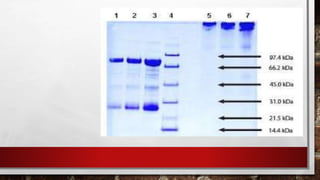
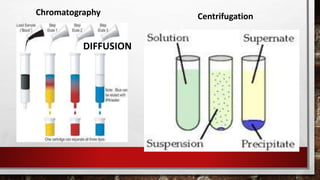
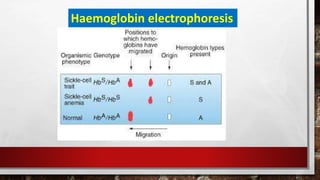
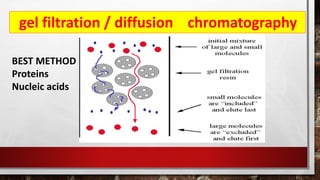
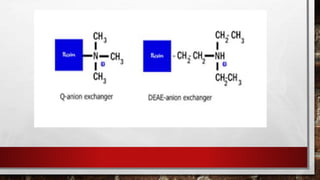
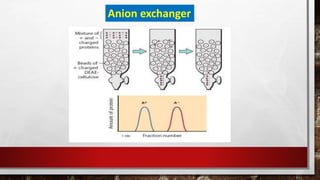
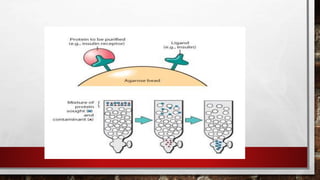
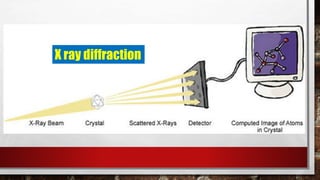
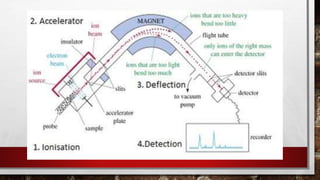
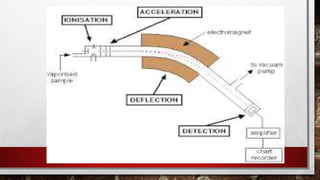
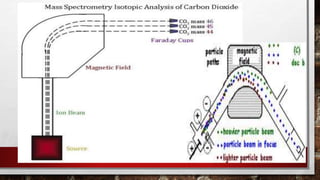
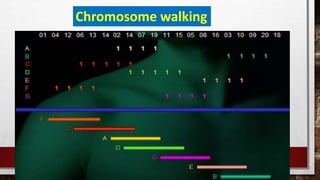
This document discusses various techniques for determining the primary, secondary, tertiary, and quaternary structures of proteins. It describes methods such as determining amino acid composition, degradation of proteins into smaller fragments, sequencing techniques like Edman degradation, and use of X-ray crystallography and NMR to analyze secondary and tertiary structures. Chromatography, electrophoresis, and centrifugation techniques are also covered for protein purification and separation.