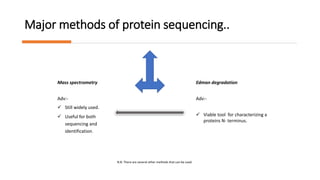
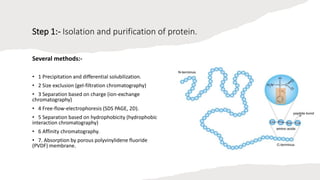
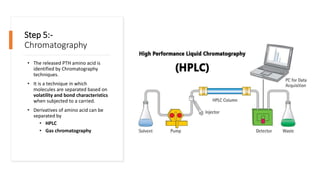
The document discusses protein sequencing, a crucial process that determines the amino acid order in proteins and aids in understanding biological functions and diseases. It outlines key methods such as mass spectrometry and Edman degradation, detailing steps like protein isolation, hydrolysis, digestion, and sequence identification. The presentation emphasizes the importance of accurate sequencing in proteomics and describes techniques used to analyze and identify protein structures.