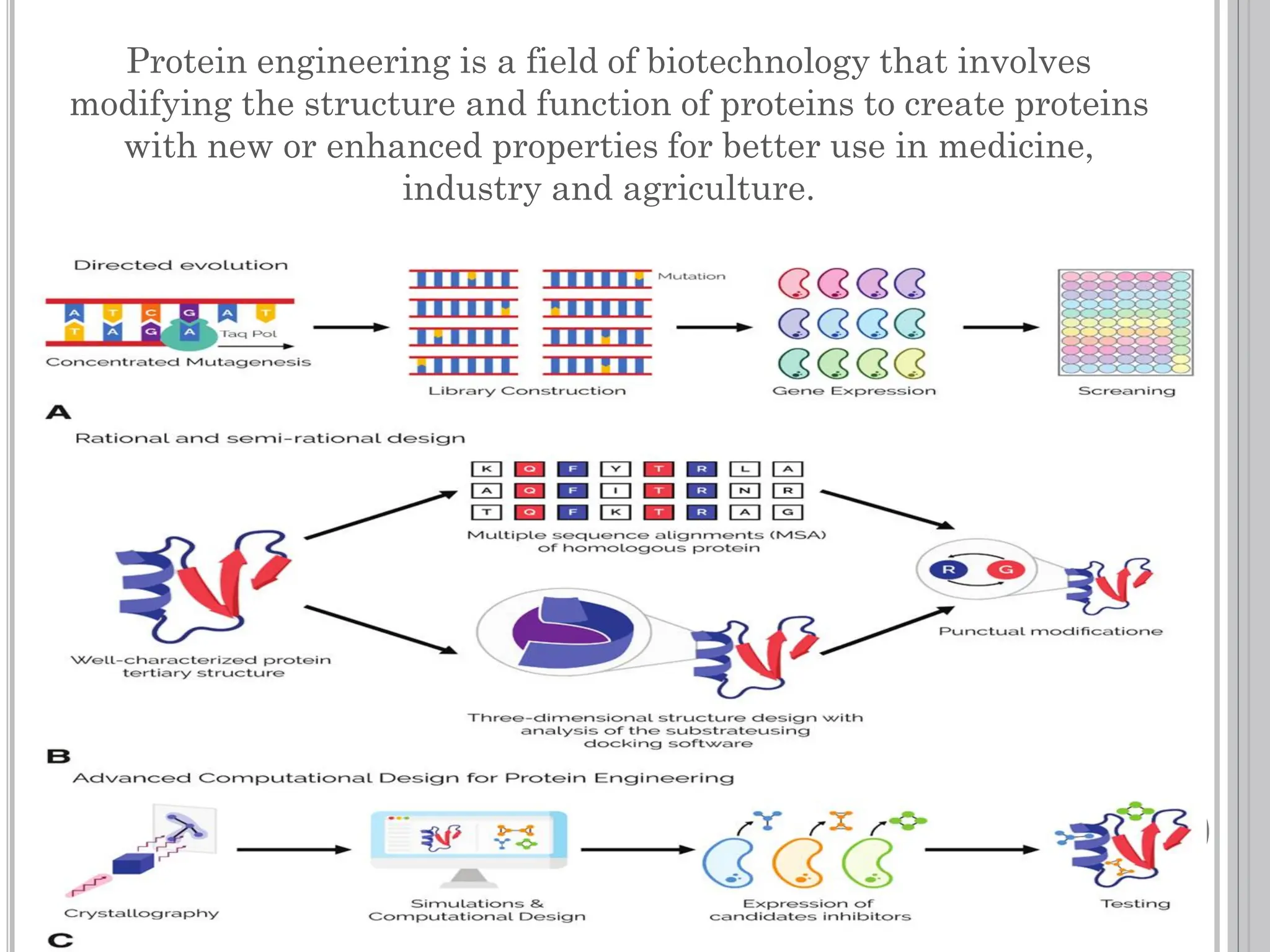
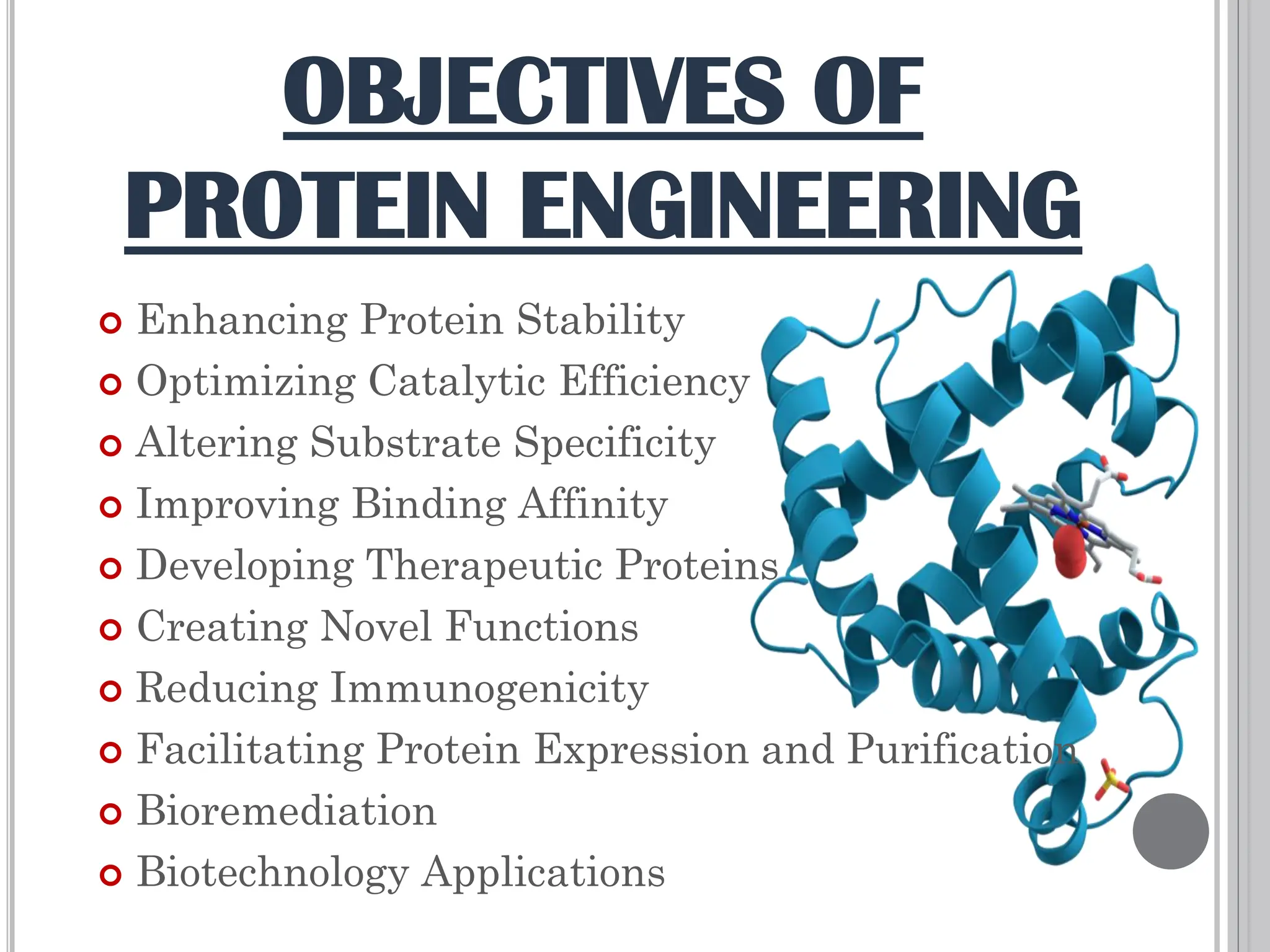
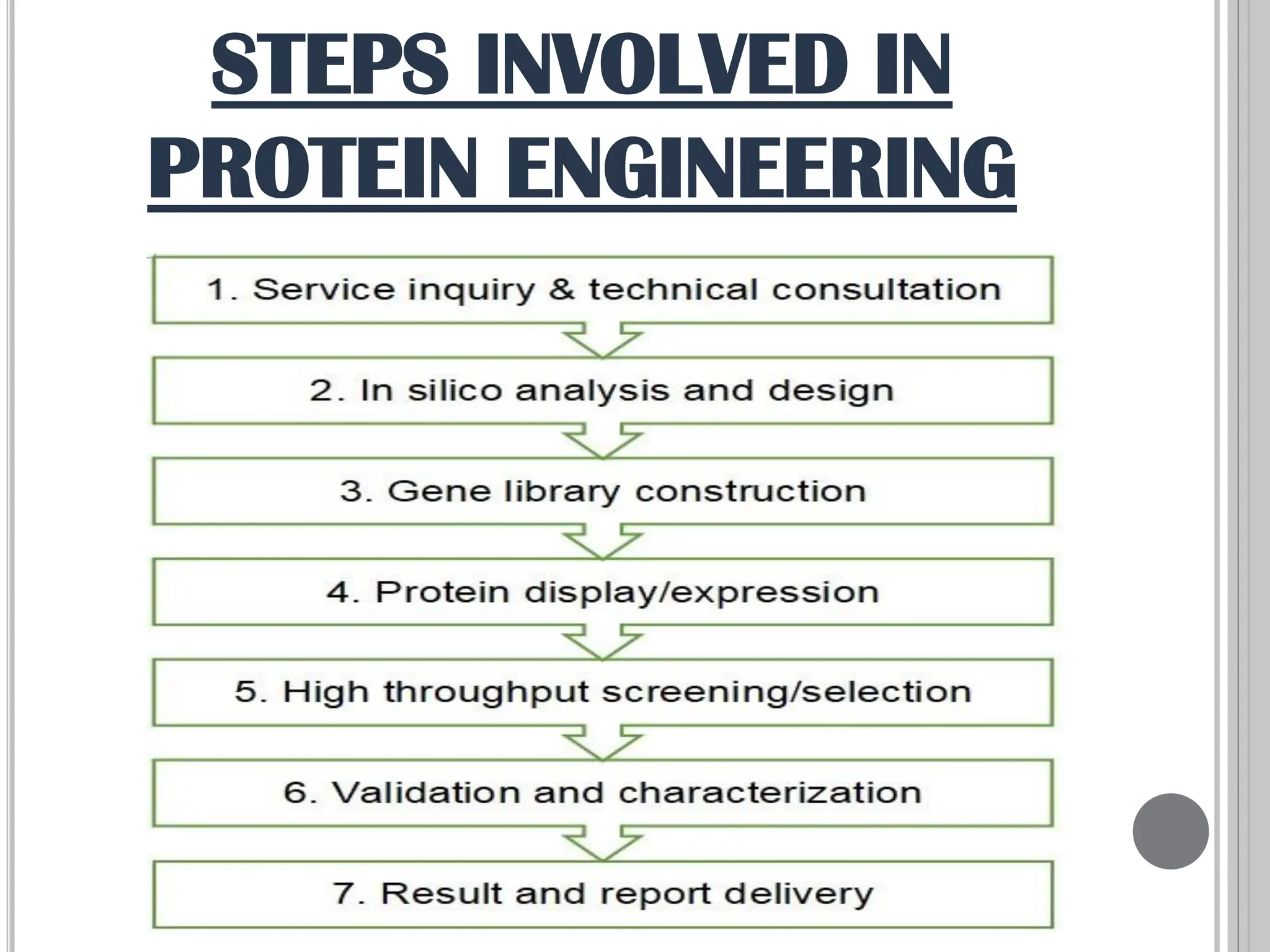
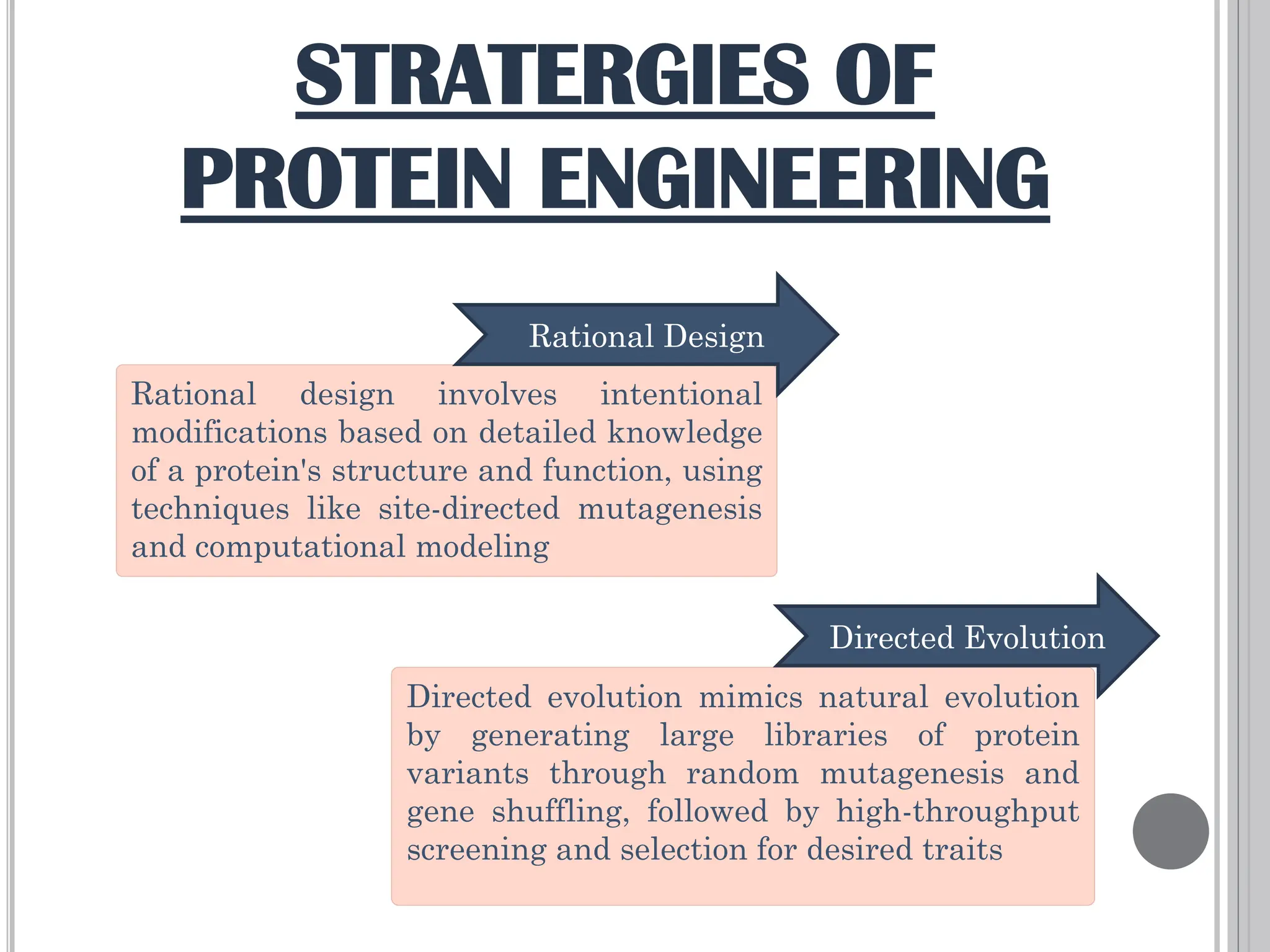
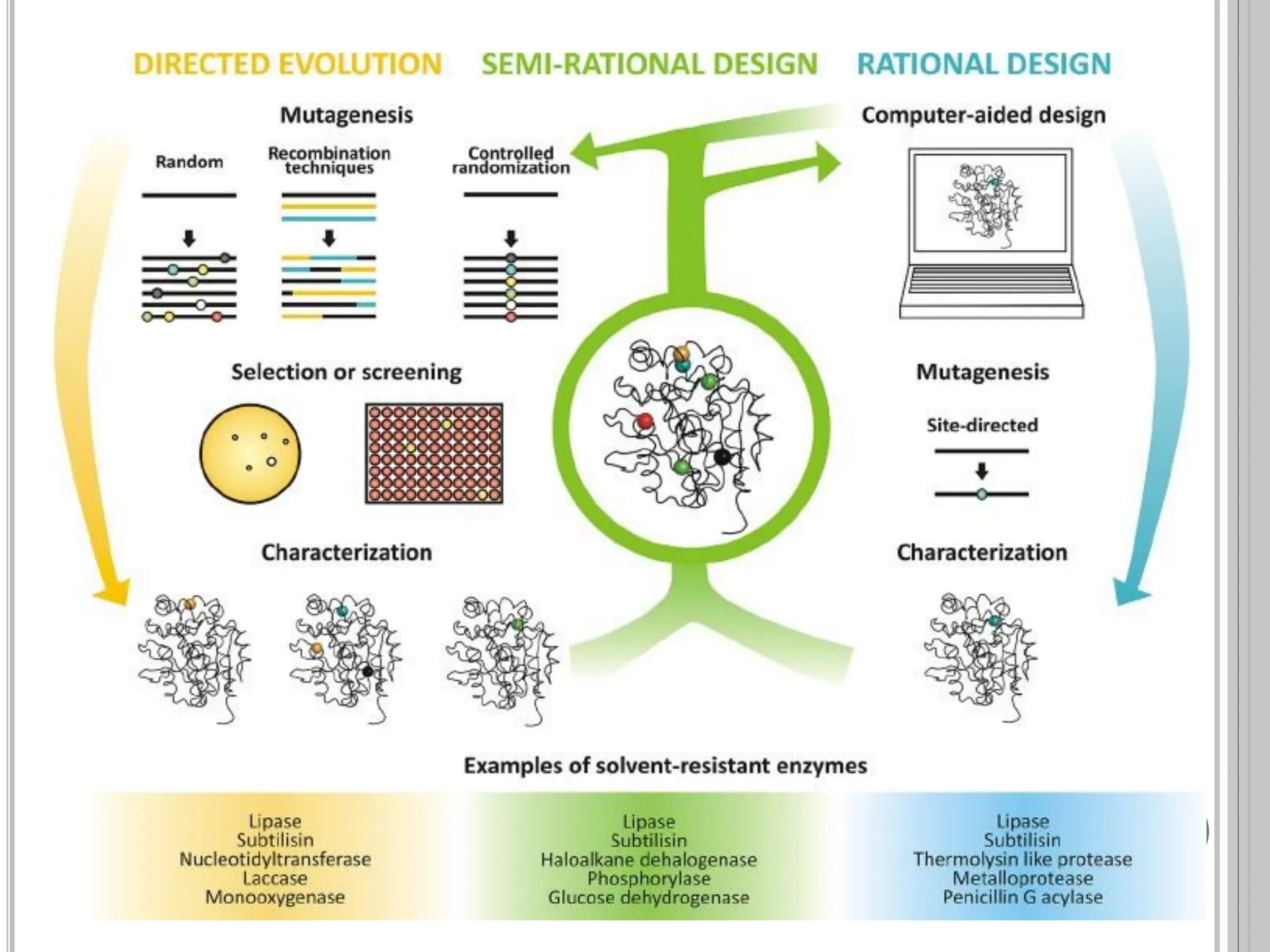
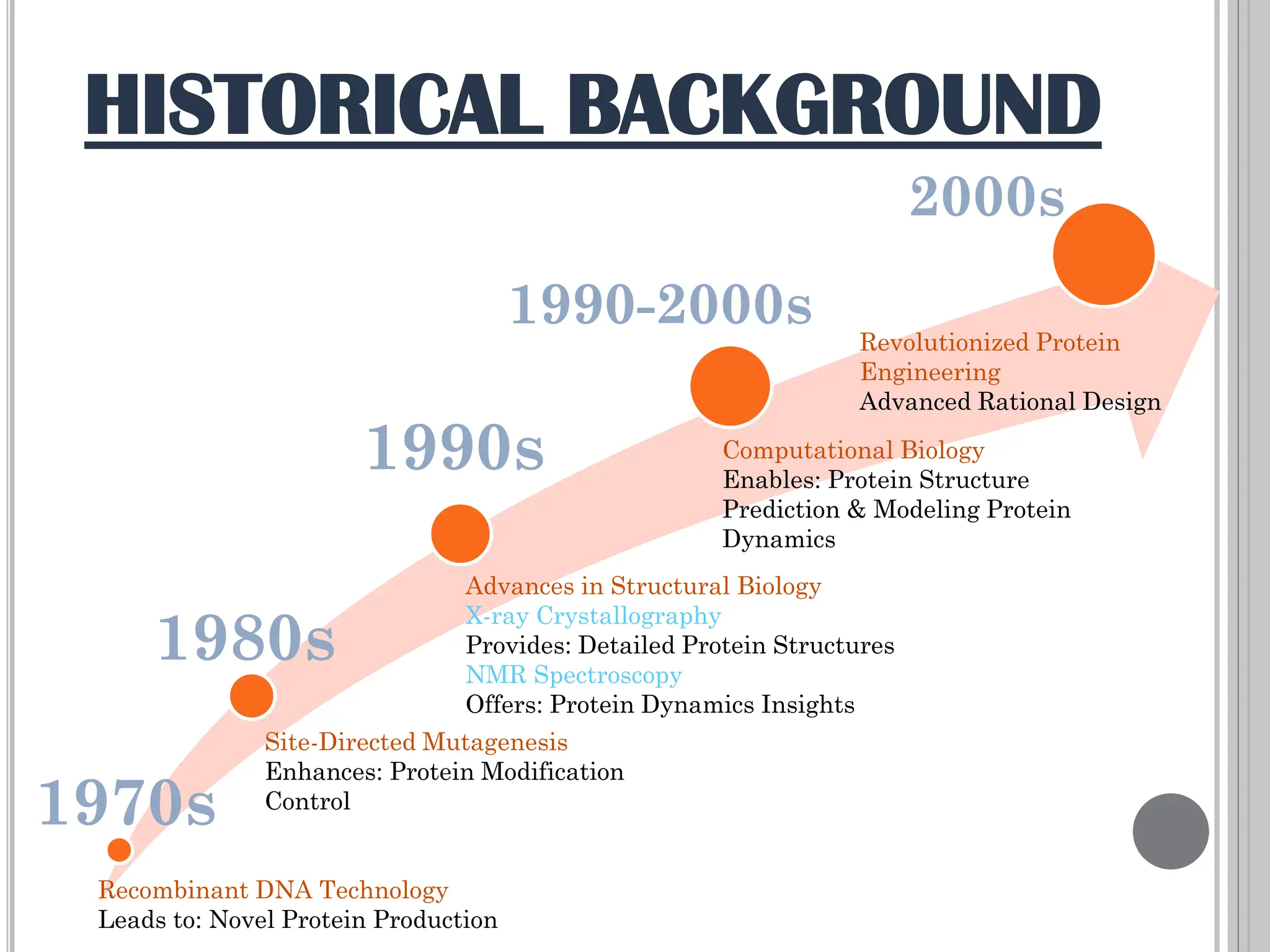
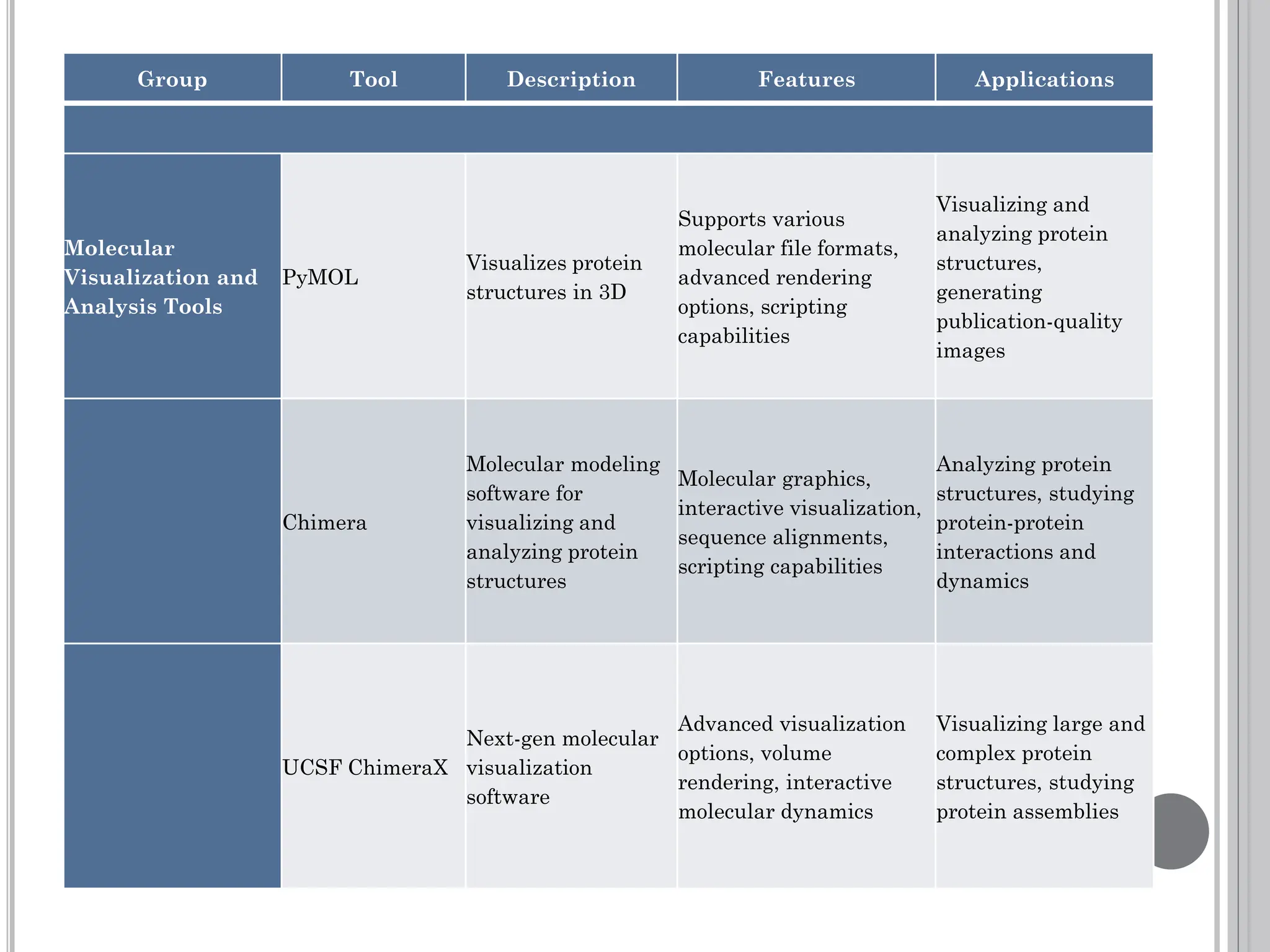
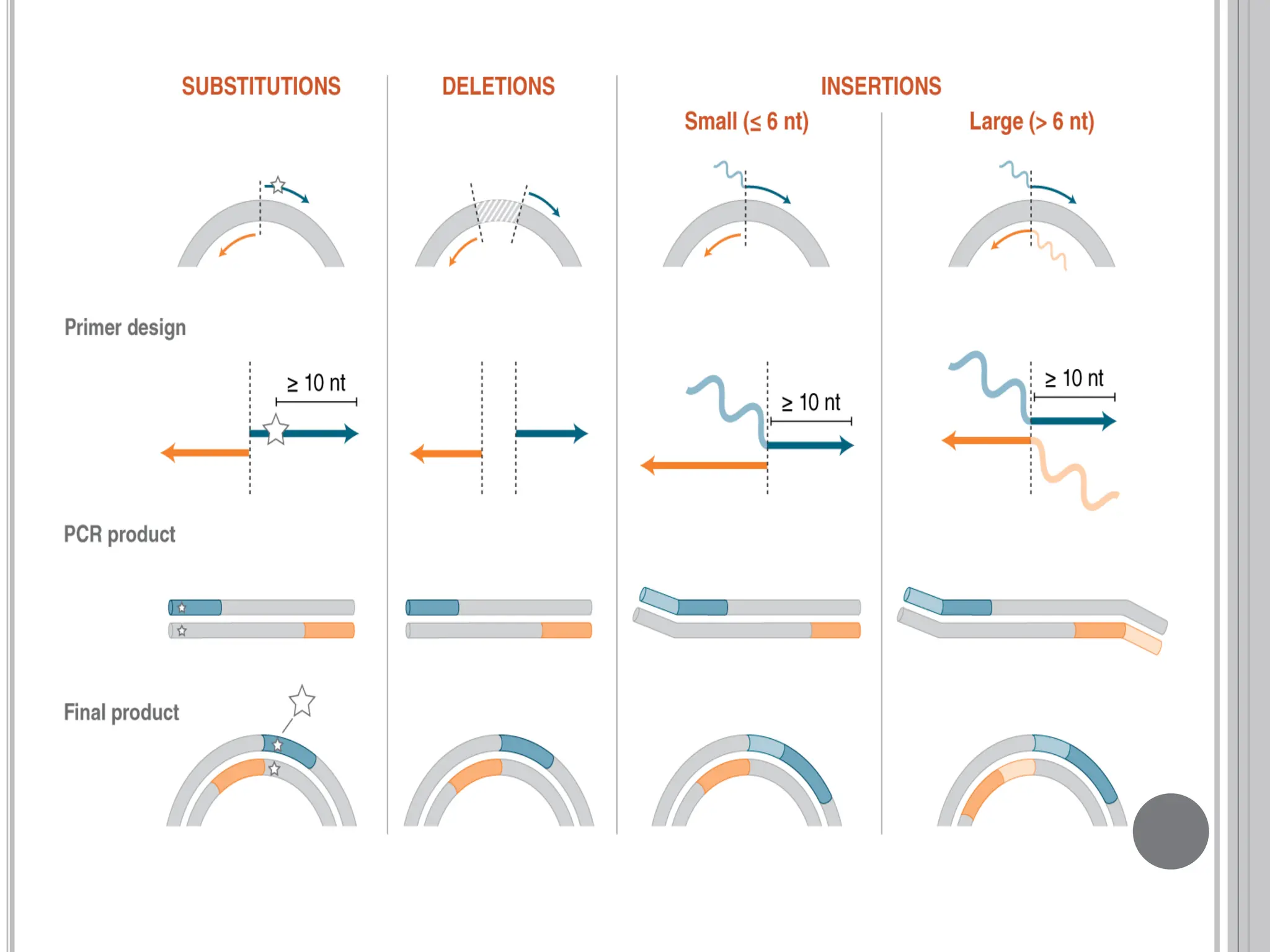
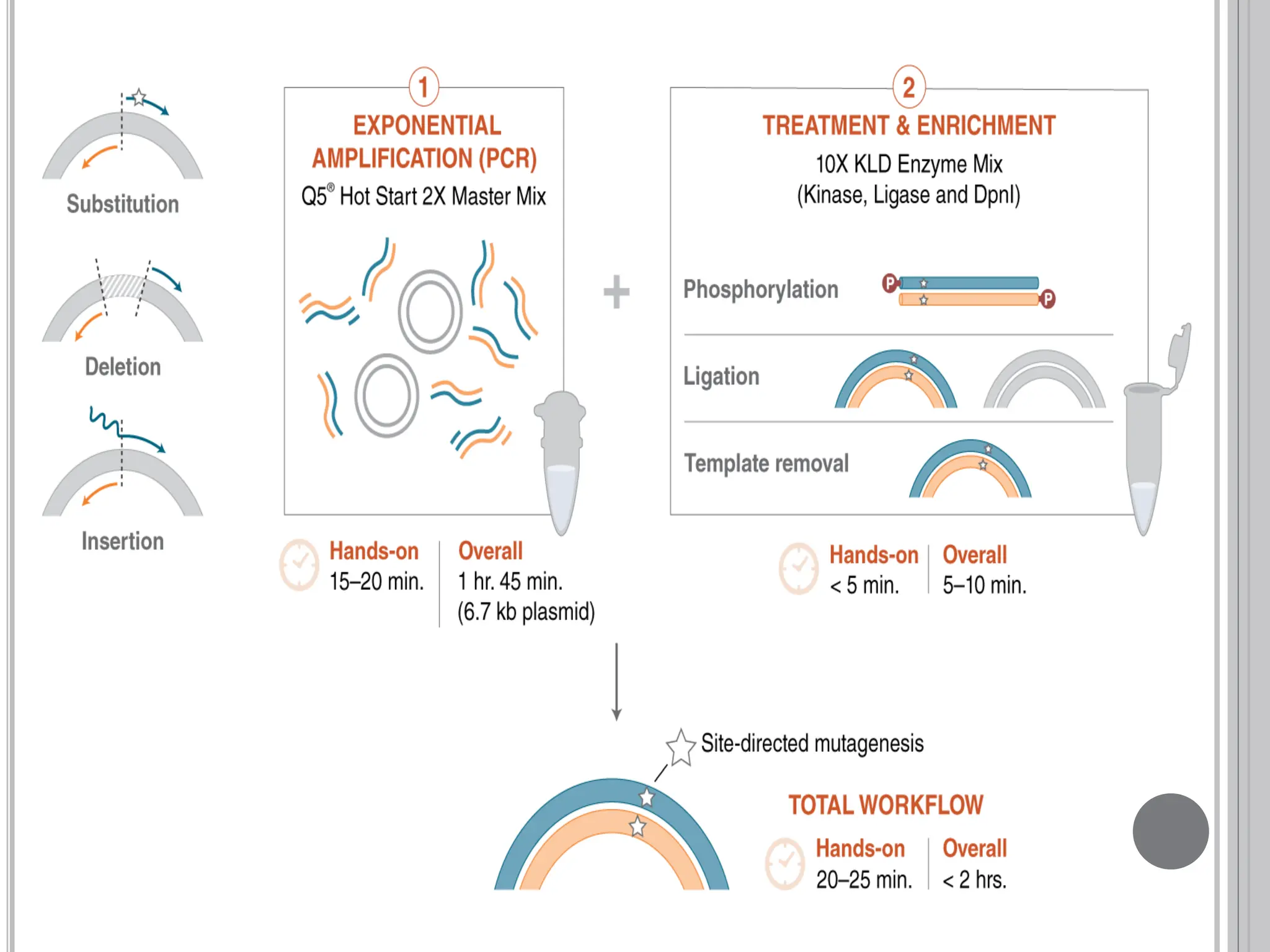
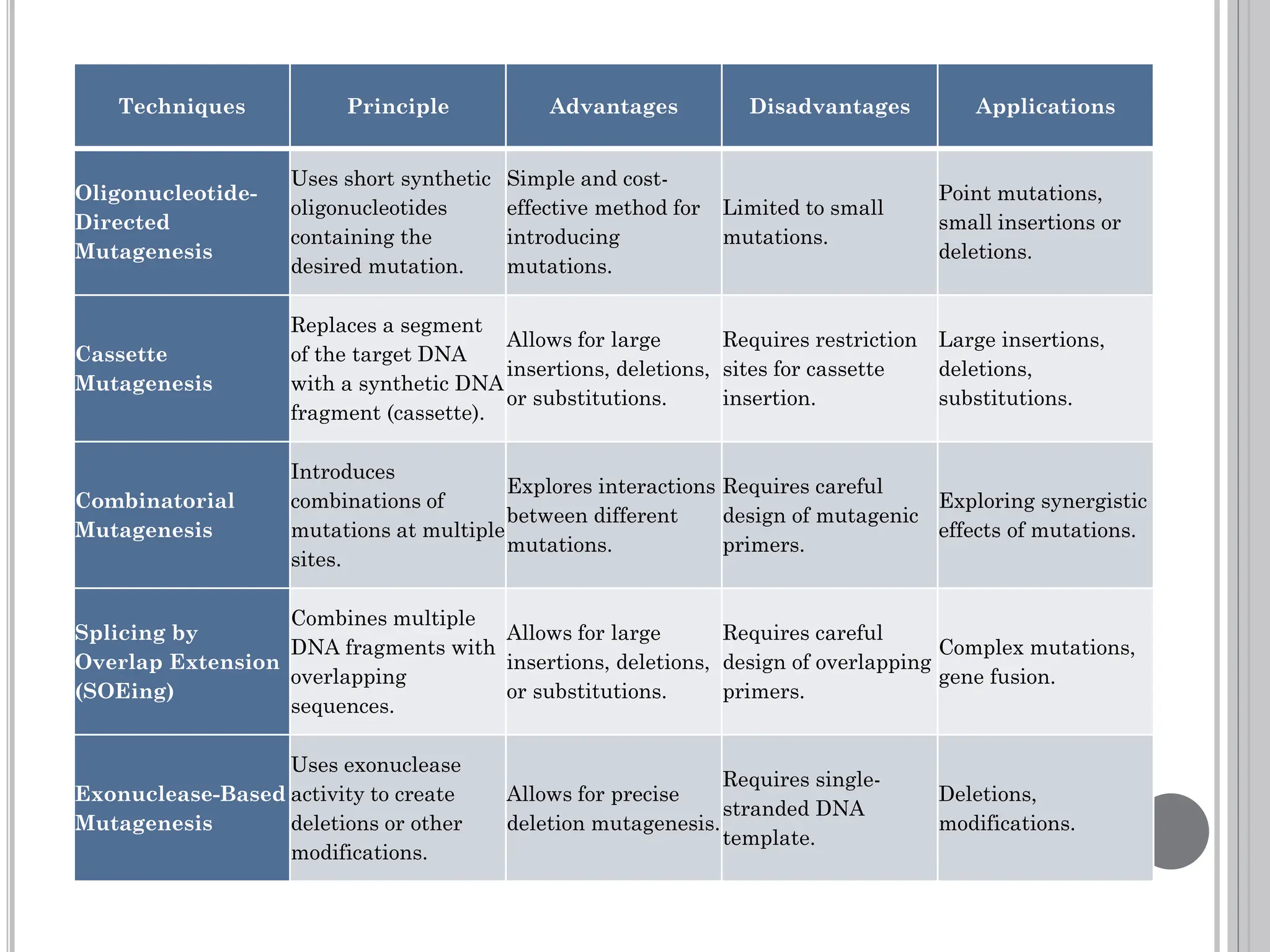
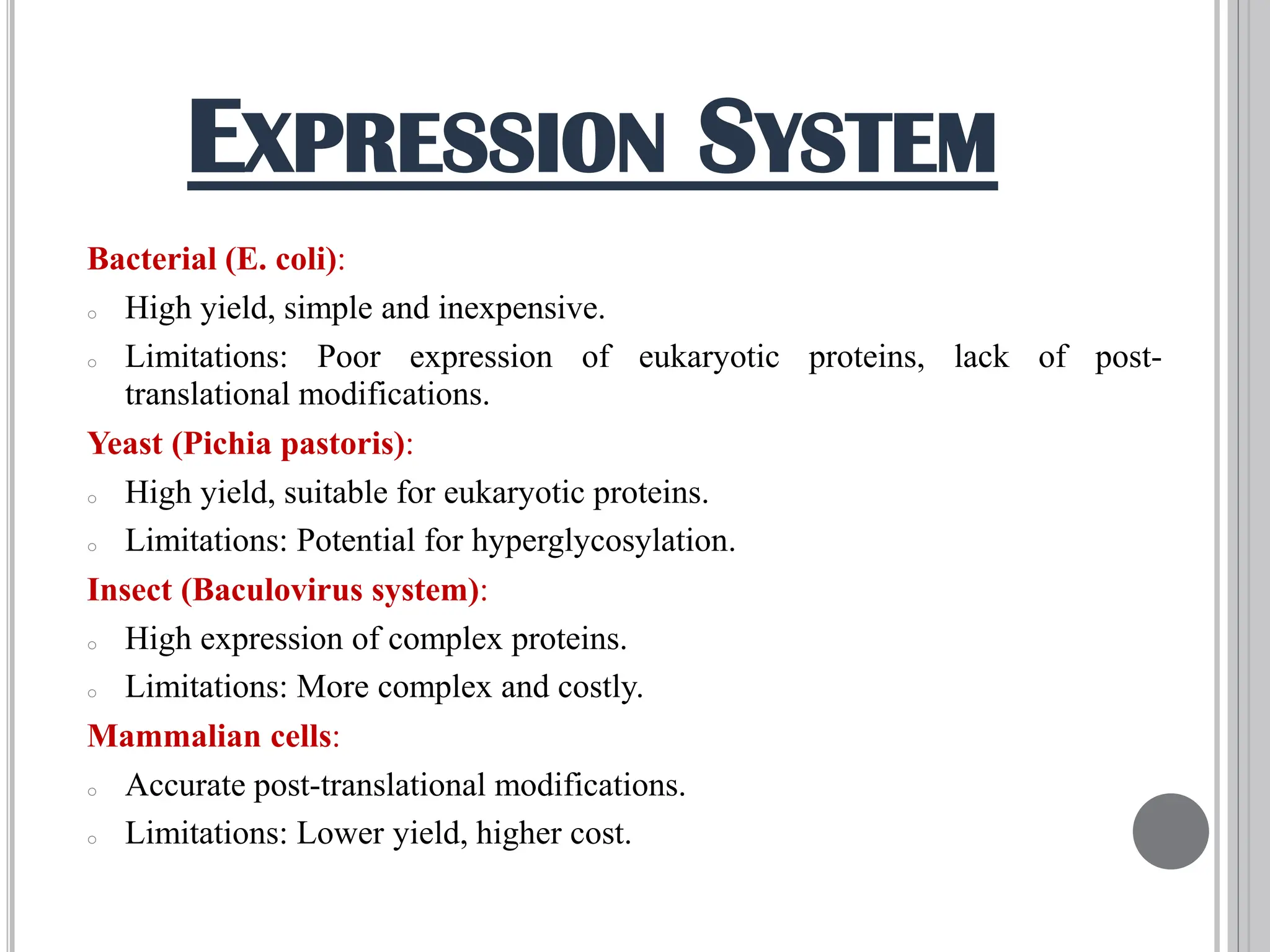
The document discusses protein engineering through rational design, detailing its objectives, strategies, and techniques. It covers the use of methods like site-directed mutagenesis and computational modeling for protein modification and highlights the importance of structural determination methods such as x-ray crystallography and NMR spectroscopy. Additionally, it describes various tools for modeling, simulation, and functional assays involved in the protein engineering process.