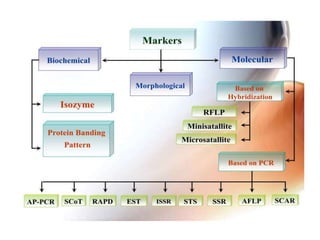
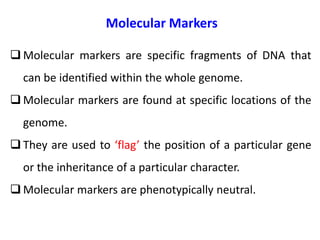
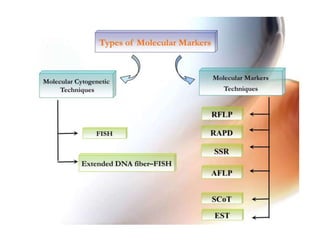
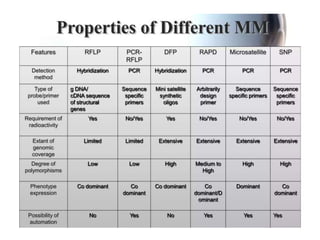
This document summarizes various molecular marker techniques used in genetics and plant breeding. It discusses restriction fragment length polymorphisms (RFLPs), randomly amplified polymorphic DNA (RAPDs), inter-simple sequence repeats (ISSRs), simple sequence repeats (SSRs), amplified fragment length polymorphisms (AFLPs), sequence-characterized amplified regions (SCARs), start codon targeted (SCoT) polymorphism analysis, and expressed sequence tags (ESTs). It also outlines applications of DNA markers such as fingerprinting, diversity studies, marker-assisted selection, genetic mapping, and gene tagging.